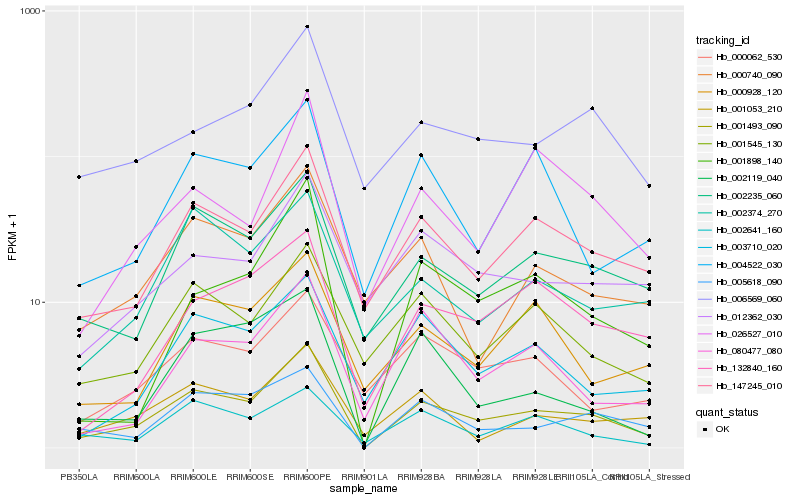
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001493_090 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_147245_010 |
0.144053636 |
- |
- |
casein kinase, putative [Ricinus communis] |
| 3 |
Hb_002235_060 |
0.1522628848 |
- |
- |
plastocyanin-like domain-containing family protein [Populus trichocarpa] |
| 4 |
Hb_005618_090 |
0.1583272656 |
transcription factor |
TF Family: AUX/IAA |
Auxin-responsive protein IAA20, putative [Ricinus communis] |
| 5 |
Hb_001898_140 |
0.1742492809 |
- |
- |
hypothetical protein AMTR_s00071p00184460 [Amborella trichopoda] |
| 6 |
Hb_001545_130 |
0.1742868032 |
- |
- |
sur2 hydroxylase/desaturase, putative [Ricinus communis] |
| 7 |
Hb_001053_210 |
0.1757571235 |
- |
- |
- |
| 8 |
Hb_006569_060 |
0.1777478061 |
- |
- |
plasma membrane intrinsic protein PIP2;3 [Hevea brasiliensis] |
| 9 |
Hb_003710_020 |
0.1791550437 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g53440 isoform X1 [Jatropha curcas] |
| 10 |
Hb_132840_160 |
0.183207542 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002641_160 |
0.1832470533 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000740_090 |
0.1834970982 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002374_270 |
0.1855961773 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_004522_030 |
0.185817967 |
- |
- |
PREDICTED: 6-phosphogluconate dehydrogenase, decarboxylating 3 [Jatropha curcas] |
| 15 |
Hb_026527_010 |
0.1862684307 |
- |
- |
PREDICTED: caffeoylshikimate esterase-like [Populus euphratica] |
| 16 |
Hb_012362_030 |
0.1866760738 |
- |
- |
PREDICTED: GPI-anchored protein LORELEI-like [Populus euphratica] |
| 17 |
Hb_080477_080 |
0.1868073194 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4 isoform X1 [Gossypium raimondii] |
| 18 |
Hb_000928_120 |
0.1876315794 |
- |
- |
PREDICTED: benzyl alcohol O-benzoyltransferase-like [Jatropha curcas] |
| 19 |
Hb_000062_530 |
0.1876805759 |
- |
- |
PREDICTED: GDP-mannose transporter GONST3-like [Populus euphratica] |
| 20 |
Hb_002119_040 |
0.1895847951 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |