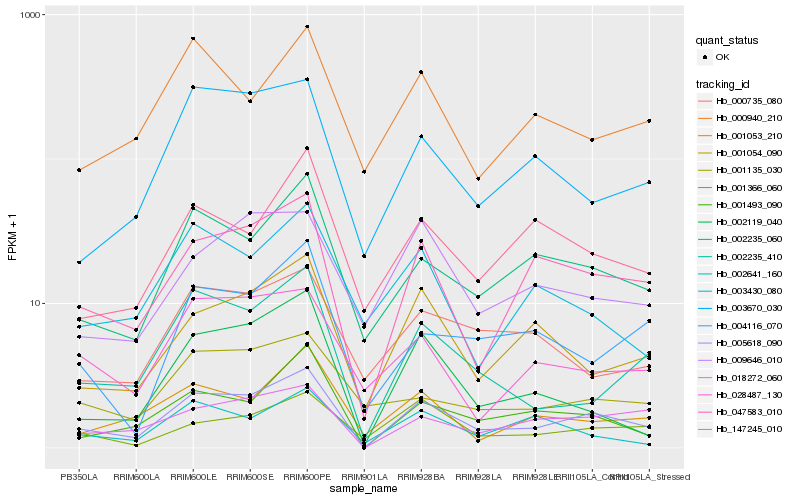
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005618_090 |
0.0 |
transcription factor |
TF Family: AUX/IAA |
Auxin-responsive protein IAA20, putative [Ricinus communis] |
| 2 |
Hb_047583_010 |
0.1363034671 |
- |
- |
hydroxyproline-rich glycoprotein [Populus trichocarpa] |
| 3 |
Hb_002235_060 |
0.1549083639 |
- |
- |
plastocyanin-like domain-containing family protein [Populus trichocarpa] |
| 4 |
Hb_001493_090 |
0.1583272656 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_018272_060 |
0.162073624 |
- |
- |
PREDICTED: mavicyanin-like [Citrus sinensis] |
| 6 |
Hb_002235_410 |
0.1666001642 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF181-like [Jatropha curcas] |
| 7 |
Hb_001054_090 |
0.1723442462 |
- |
- |
Potassium channel SKOR, putative [Ricinus communis] |
| 8 |
Hb_003670_030 |
0.1812210297 |
- |
- |
PREDICTED: probable calcium-binding protein CML35 [Jatropha curcas] |
| 9 |
Hb_002119_040 |
0.1844600741 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 10 |
Hb_004116_070 |
0.1869660864 |
- |
- |
PREDICTED: uncharacterized protein LOC105646325 [Jatropha curcas] |
| 11 |
Hb_147245_010 |
0.1879502 |
- |
- |
casein kinase, putative [Ricinus communis] |
| 12 |
Hb_001053_210 |
0.1885436133 |
- |
- |
- |
| 13 |
Hb_028487_130 |
0.191068347 |
- |
- |
kinase family protein [Populus trichocarpa] |
| 14 |
Hb_001366_060 |
0.1912493089 |
- |
- |
- |
| 15 |
Hb_000735_080 |
0.1914442634 |
- |
- |
PREDICTED: probable xyloglucan glycosyltransferase 6 [Jatropha curcas] |
| 16 |
Hb_009646_010 |
0.1947456915 |
- |
- |
PREDICTED: hyoscyamine 6-dioxygenase-like [Jatropha curcas] |
| 17 |
Hb_001135_030 |
0.1951391282 |
- |
- |
PREDICTED: interaptin-like [Jatropha curcas] |
| 18 |
Hb_000940_210 |
0.195715293 |
- |
- |
cyclophilin [Hevea brasiliensis] |
| 19 |
Hb_003430_080 |
0.2007958394 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 20 |
Hb_002641_160 |
0.2010180789 |
- |
- |
conserved hypothetical protein [Ricinus communis] |