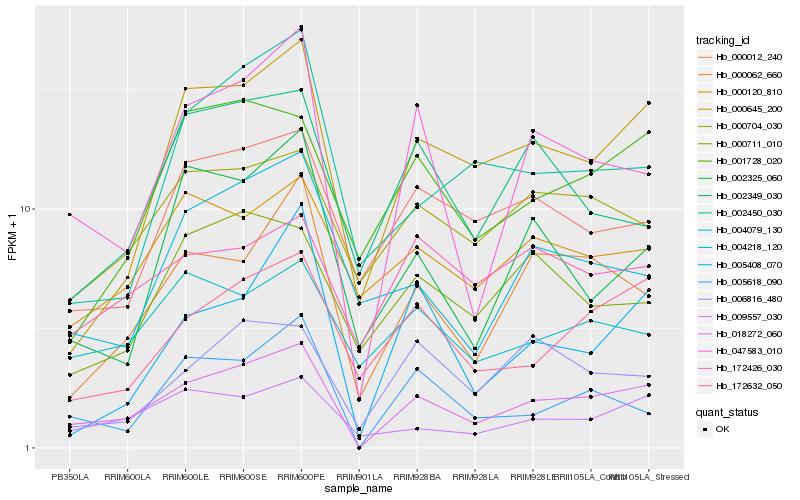
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_018272_060 |
0.0 |
- |
- |
PREDICTED: mavicyanin-like [Citrus sinensis] |
| 2 |
Hb_047583_010 |
0.1336787037 |
- |
- |
hydroxyproline-rich glycoprotein [Populus trichocarpa] |
| 3 |
Hb_000645_200 |
0.1526933917 |
- |
- |
PREDICTED: GTP-binding protein SAR1A [Jatropha curcas] |
| 4 |
Hb_000062_660 |
0.1558804638 |
- |
- |
hypothetical protein POPTR_0009s03470g [Populus trichocarpa] |
| 5 |
Hb_005618_090 |
0.162073624 |
transcription factor |
TF Family: AUX/IAA |
Auxin-responsive protein IAA20, putative [Ricinus communis] |
| 6 |
Hb_009557_030 |
0.1659868935 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein ATHB-52-like [Jatropha curcas] |
| 7 |
Hb_172632_050 |
0.1716014923 |
- |
- |
PREDICTED: uncharacterized protein LOC105646134 [Jatropha curcas] |
| 8 |
Hb_001728_020 |
0.1802890045 |
- |
- |
PREDICTED: trans-1,2-dihydrobenzene-1,2-diol dehydrogenase [Jatropha curcas] |
| 9 |
Hb_000012_240 |
0.1810105549 |
- |
- |
PREDICTED: ferrochelatase-2, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000704_030 |
0.1825831298 |
- |
- |
PREDICTED: probable aquaporin SIP2-1 [Cicer arietinum] |
| 11 |
Hb_006816_480 |
0.1852151018 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 3 isoform X1 [Jatropha curcas] |
| 12 |
Hb_002325_060 |
0.1855950688 |
- |
- |
PREDICTED: ADP-ribosylation factor-like protein 8B-like [Glycine max] |
| 13 |
Hb_004218_120 |
0.1859572014 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase SINAT5-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_000120_810 |
0.1867550192 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 15 |
Hb_005408_070 |
0.186903513 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP4 [Jatropha curcas] |
| 16 |
Hb_004079_130 |
0.1896882058 |
- |
- |
PREDICTED: uncharacterized protein LOC105637708 [Jatropha curcas] |
| 17 |
Hb_002349_030 |
0.1902163115 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000711_010 |
0.1902993625 |
transcription factor |
TF Family: Tify |
PREDICTED: protein TIFY 6B isoform X1 [Jatropha curcas] |
| 19 |
Hb_002450_030 |
0.1903213415 |
- |
- |
ATPP2-A13, putative [Ricinus communis] |
| 20 |
Hb_172426_030 |
0.1920511106 |
- |
- |
acetylglucosaminyltransferase, putative [Ricinus communis] |