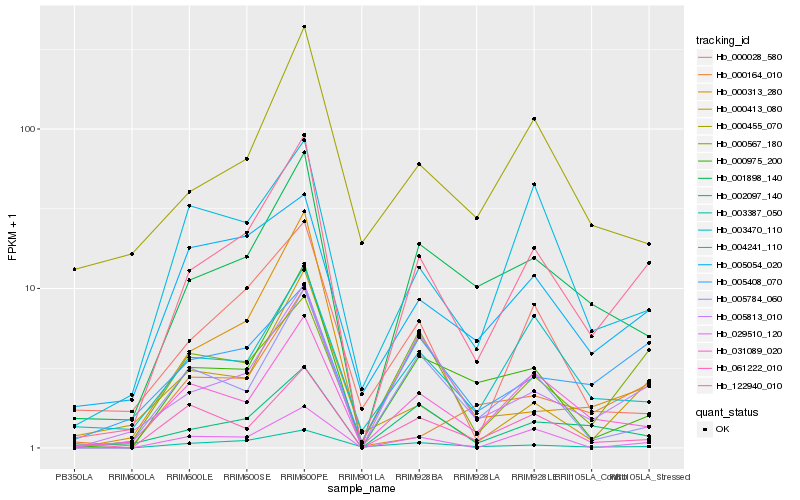
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_122940_010 |
0.0 |
- |
- |
PREDICTED: 2-isopropylmalate synthase 2, chloroplastic-like [Populus euphratica] |
| 2 |
Hb_001898_140 |
0.1685550476 |
- |
- |
hypothetical protein AMTR_s00071p00184460 [Amborella trichopoda] |
| 3 |
Hb_005813_010 |
0.1690892868 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 4 |
Hb_031089_020 |
0.1725901768 |
- |
- |
Nodulation receptor kinase precursor, putative [Ricinus communis] |
| 5 |
Hb_000975_200 |
0.1802655205 |
- |
- |
PREDICTED: uncharacterized protein LOC105628231 isoform X2 [Jatropha curcas] |
| 6 |
Hb_003387_050 |
0.1840899788 |
- |
- |
PREDICTED: uncharacterized protein LOC105639324 [Jatropha curcas] |
| 7 |
Hb_002097_140 |
0.187172642 |
- |
- |
CHY zinc finger family protein, expressed [Oryza sativa Japonica Group] |
| 8 |
Hb_029510_120 |
0.1918876899 |
- |
- |
PREDICTED: protein PLANT CADMIUM RESISTANCE 8-like [Jatropha curcas] |
| 9 |
Hb_000313_280 |
0.1951471177 |
- |
- |
hypothetical protein PRUPE_ppa014856mg, partial [Prunus persica] |
| 10 |
Hb_005784_060 |
0.1952315168 |
- |
- |
PREDICTED: U-box domain-containing protein 28 [Jatropha curcas] |
| 11 |
Hb_003470_110 |
0.1958149964 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 12 |
Hb_000413_080 |
0.2001150069 |
- |
- |
- |
| 13 |
Hb_005408_070 |
0.2028940404 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP4 [Jatropha curcas] |
| 14 |
Hb_000164_010 |
0.2038464377 |
- |
- |
small heat-shock protein, putative [Ricinus communis] |
| 15 |
Hb_005054_020 |
0.2088545868 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g11320 [Jatropha curcas] |
| 16 |
Hb_000028_580 |
0.2103232537 |
- |
- |
PREDICTED: glutamate decarboxylase 5-like [Jatropha curcas] |
| 17 |
Hb_000567_180 |
0.2103373147 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF1.5 [Jatropha curcas] |
| 18 |
Hb_000455_070 |
0.2121701812 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: carboxymethylenebutenolidase homolog [Jatropha curcas] |
| 19 |
Hb_061222_010 |
0.2126972019 |
- |
- |
hypothetical protein RCOM_0012650 [Ricinus communis] |
| 20 |
Hb_004241_110 |
0.2129788905 |
- |
- |
Minor allergen Alt a, putative [Ricinus communis] |