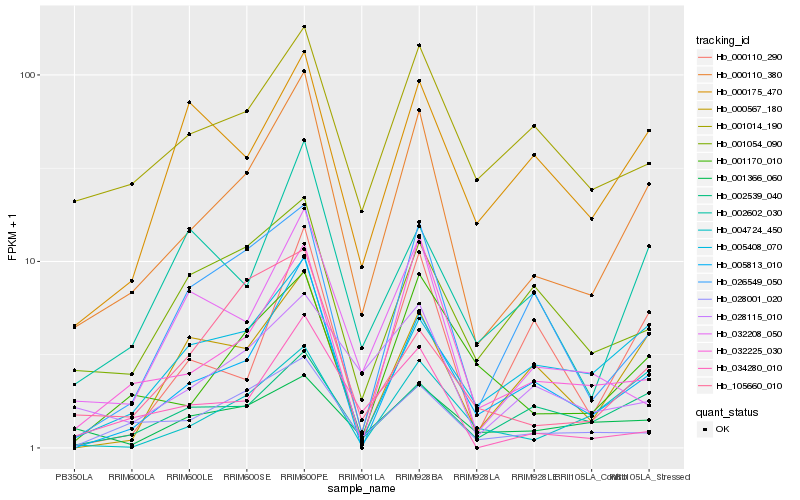
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000110_380 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642019 [Jatropha curcas] |
| 2 |
Hb_005813_010 |
0.1643268054 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 3 |
Hb_032225_030 |
0.1737273537 |
- |
- |
PREDICTED: leucine-rich repeat extensin-like protein 2 [Jatropha curcas] |
| 4 |
Hb_028001_020 |
0.1768224892 |
- |
- |
PREDICTED: lysM domain receptor-like kinase 4 [Jatropha curcas] |
| 5 |
Hb_002539_040 |
0.1839090635 |
- |
- |
SPFH domain-containing protein 2 precursor, putative [Ricinus communis] |
| 6 |
Hb_001170_010 |
0.1861788819 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 7 |
Hb_005408_070 |
0.186324706 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP4 [Jatropha curcas] |
| 8 |
Hb_002602_030 |
0.1951813606 |
- |
- |
PREDICTED: chaperone protein dnaJ 11, chloroplastic [Jatropha curcas] |
| 9 |
Hb_001014_190 |
0.1954633936 |
- |
- |
PREDICTED: uncharacterized protein LOC105647294 [Jatropha curcas] |
| 10 |
Hb_034280_010 |
0.1955968769 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 11 |
Hb_028115_010 |
0.1958691479 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000567_180 |
0.2012972046 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF1.5 [Jatropha curcas] |
| 13 |
Hb_026549_050 |
0.2032535526 |
- |
- |
PREDICTED: uncharacterized protein LOC105636246 isoform X2 [Jatropha curcas] |
| 14 |
Hb_032208_050 |
0.2047971176 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 4 isoform X3 [Jatropha curcas] |
| 15 |
Hb_001366_060 |
0.2084142206 |
- |
- |
- |
| 16 |
Hb_001054_090 |
0.209374242 |
- |
- |
Potassium channel SKOR, putative [Ricinus communis] |
| 17 |
Hb_004724_450 |
0.2105291623 |
- |
- |
PREDICTED: uncharacterized protein LOC105647069 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000175_470 |
0.2112562553 |
- |
- |
lactoylglutathione lyase, putative [Ricinus communis] |
| 19 |
Hb_000110_290 |
0.2120795085 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_105660_010 |
0.2152176462 |
- |
- |
PREDICTED: potassium channel AKT6-like [Jatropha curcas] |