| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
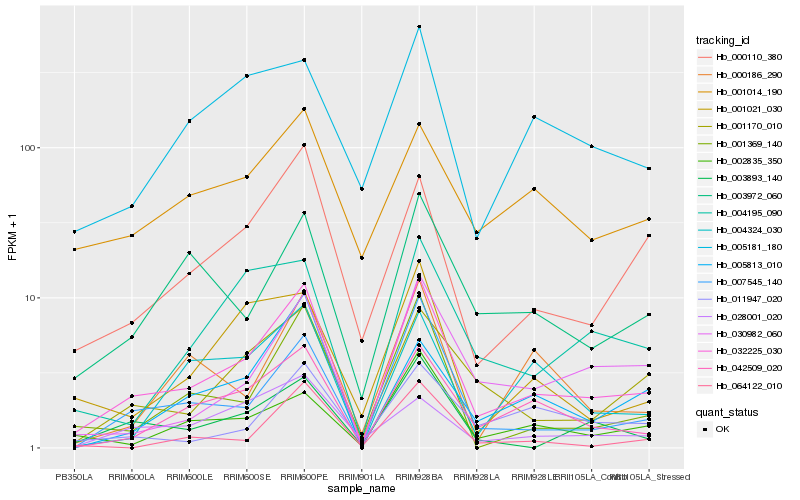
Hb_032225_030 |
0.0 |
- |
- |
PREDICTED: leucine-rich repeat extensin-like protein 2 [Jatropha curcas] |
| 2 |
Hb_042509_020 |
0.1526128881 |
- |
- |
hypothetical protein JCGZ_19555 [Jatropha curcas] |
| 3 |
Hb_007545_140 |
0.1558882679 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105141503 [Populus euphratica] |
| 4 |
Hb_001369_140 |
0.1725698785 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001170_010 |
0.1726248333 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 6 |
Hb_000110_380 |
0.1737273537 |
- |
- |
PREDICTED: uncharacterized protein LOC105642019 [Jatropha curcas] |
| 7 |
Hb_030982_060 |
0.1809122501 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_003893_140 |
0.1835353749 |
- |
- |
palmitoyl-acyl carrier protein thioesterase [Ricinus communis] |
| 9 |
Hb_003972_060 |
0.1886480533 |
- |
- |
PREDICTED: UDP-arabinopyranose mutase 3 [Populus euphratica] |
| 10 |
Hb_002835_350 |
0.196863852 |
- |
- |
PREDICTED: uncharacterized protein LOC105640319 [Jatropha curcas] |
| 11 |
Hb_011947_020 |
0.198186318 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 12 |
Hb_004195_090 |
0.1999148698 |
- |
- |
PREDICTED: probable mitochondrial chaperone bcs1 [Jatropha curcas] |
| 13 |
Hb_005813_010 |
0.2016715745 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 14 |
Hb_001021_030 |
0.2024522564 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 15 |
Hb_000186_290 |
0.2042252931 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 8 isoform X1 [Jatropha curcas] |
| 16 |
Hb_064122_010 |
0.2046551175 |
- |
- |
PREDICTED: uncharacterized protein LOC105631858 isoform X2 [Jatropha curcas] |
| 17 |
Hb_028001_020 |
0.2064202627 |
- |
- |
PREDICTED: lysM domain receptor-like kinase 4 [Jatropha curcas] |
| 18 |
Hb_004324_030 |
0.2075474417 |
- |
- |
PREDICTED: probable protein phosphatase 2C 8 [Jatropha curcas] |
| 19 |
Hb_001014_190 |
0.2083300341 |
- |
- |
PREDICTED: uncharacterized protein LOC105647294 [Jatropha curcas] |
| 20 |
Hb_005181_180 |
0.2088951495 |
- |
- |
conserved hypothetical protein [Ricinus communis] |