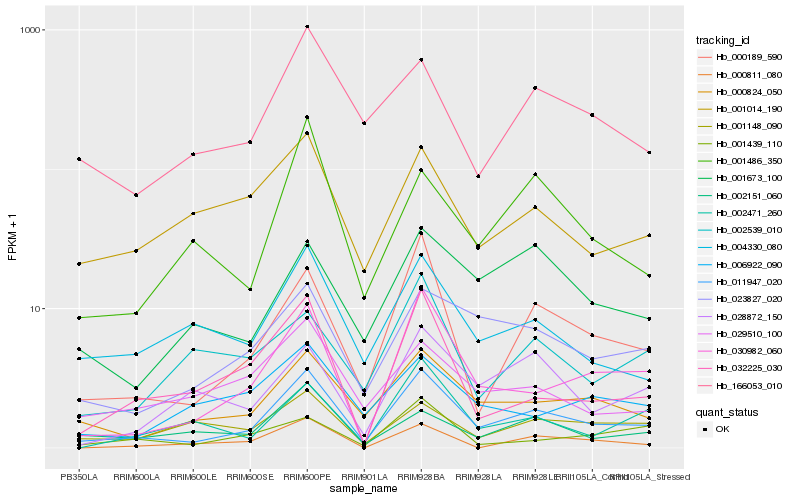
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011947_020 |
0.0 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 2 |
Hb_000189_590 |
0.1490327476 |
- |
- |
PREDICTED: cysteine synthase [Jatropha curcas] |
| 3 |
Hb_030982_060 |
0.150740893 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000824_050 |
0.1682246047 |
- |
- |
PREDICTED: nicotinate phosphoribosyltransferase 1 [Jatropha curcas] |
| 5 |
Hb_023827_020 |
0.187326691 |
- |
- |
beta-1,3-glucanase 1 [Ziziphus jujuba] |
| 6 |
Hb_032225_030 |
0.198186318 |
- |
- |
PREDICTED: leucine-rich repeat extensin-like protein 2 [Jatropha curcas] |
| 7 |
Hb_004330_080 |
0.1995743953 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 8 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001486_350 |
0.2025075202 |
- |
- |
5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase, putative [Ricinus communis] |
| 9 |
Hb_166053_010 |
0.2029649734 |
- |
- |
glyceraldehyde-3-phosphate dehydrogenase, partial [Vernicia fordii] |
| 10 |
Hb_002151_060 |
0.2089511999 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit rpc4 [Jatropha curcas] |
| 11 |
Hb_001014_190 |
0.2092091735 |
- |
- |
PREDICTED: uncharacterized protein LOC105647294 [Jatropha curcas] |
| 12 |
Hb_028872_150 |
0.2098051596 |
- |
- |
PREDICTED: uncharacterized protein LOC105137493 [Populus euphratica] |
| 13 |
Hb_001673_100 |
0.2104294207 |
- |
- |
PREDICTED: putative phosphatidylglycerol/phosphatidylinositol transfer protein DDB_G0282179 [Jatropha curcas] |
| 14 |
Hb_029510_100 |
0.2129658892 |
- |
- |
scab resistance family protein [Populus trichocarpa] |
| 15 |
Hb_002471_260 |
0.2142222066 |
transcription factor |
TF Family: PLATZ |
PREDICTED: uncharacterized protein LOC105634154 [Jatropha curcas] |
| 16 |
Hb_001148_090 |
0.2199459441 |
- |
- |
hypothetical protein JCGZ_14711 [Jatropha curcas] |
| 17 |
Hb_001439_110 |
0.2212958628 |
transcription factor |
TF Family: ERF |
PREDICTED: uncharacterized protein LOC105133481 [Populus euphratica] |
| 18 |
Hb_006922_090 |
0.2250107166 |
- |
- |
PREDICTED: interaptin [Jatropha curcas] |
| 19 |
Hb_000811_080 |
0.225575476 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002539_010 |
0.2256240245 |
- |
- |
hypothetical protein JCGZ_15711 [Jatropha curcas] |