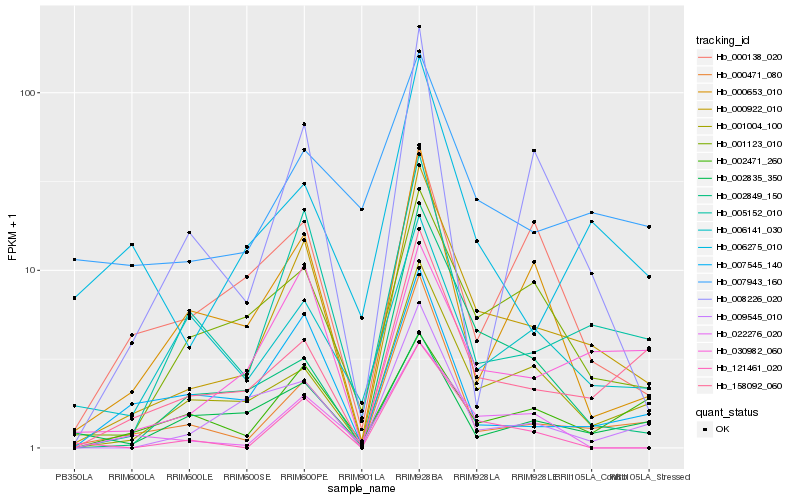
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000922_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_11488 [Jatropha curcas] |
| 2 |
Hb_005152_010 |
0.1711158904 |
- |
- |
hypothetical protein JCGZ_22048 [Jatropha curcas] |
| 3 |
Hb_001004_100 |
0.1829835696 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 4 |
Hb_158092_060 |
0.1932061765 |
- |
- |
aspartate semialdehyde dehydrogenase, putative [Ricinus communis] |
| 5 |
Hb_000471_080 |
0.1944336889 |
- |
- |
PREDICTED: probable mitochondrial chaperone BCS1-A [Jatropha curcas] |
| 6 |
Hb_001123_010 |
0.1977005802 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_030982_060 |
0.1990145954 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_009545_010 |
0.2123199767 |
transcription factor |
TF Family: GNAT |
Acetyltransferase [Medicago truncatula] |
| 9 |
Hb_000653_010 |
0.2147520118 |
- |
- |
hypothetical protein CICLE_v10033238mg [Citrus clementina] |
| 10 |
Hb_007545_140 |
0.2168222493 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105141503 [Populus euphratica] |
| 11 |
Hb_002849_150 |
0.2177143768 |
- |
- |
hypothetical protein JCGZ_14367 [Jatropha curcas] |
| 12 |
Hb_022276_020 |
0.2247389424 |
- |
- |
- |
| 13 |
Hb_121461_020 |
0.2319267132 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 14 |
Hb_008226_020 |
0.2333324359 |
- |
- |
hypothetical protein JCGZ_03321 [Jatropha curcas] |
| 15 |
Hb_006275_010 |
0.2349835884 |
- |
- |
PREDICTED: ribulose bisphosphate carboxylase small chain clone 512-like [Jatropha curcas] |
| 16 |
Hb_002471_260 |
0.2387730027 |
transcription factor |
TF Family: PLATZ |
PREDICTED: uncharacterized protein LOC105634154 [Jatropha curcas] |
| 17 |
Hb_006141_030 |
0.2402979718 |
- |
- |
- |
| 18 |
Hb_002835_350 |
0.2414218734 |
- |
- |
PREDICTED: uncharacterized protein LOC105640319 [Jatropha curcas] |
| 19 |
Hb_000138_020 |
0.2417291628 |
- |
- |
PREDICTED: peroxidase A2-like [Jatropha curcas] |
| 20 |
Hb_007943_160 |
0.2417724837 |
- |
- |
PREDICTED: aspartate aminotransferase, cytoplasmic [Jatropha curcas] |