| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000140_190 |
0.0 |
- |
- |
acc synthase, putative [Ricinus communis] |
| 2 |
Hb_000748_070 |
0.141224898 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0005s22480g [Populus trichocarpa] |
| 3 |
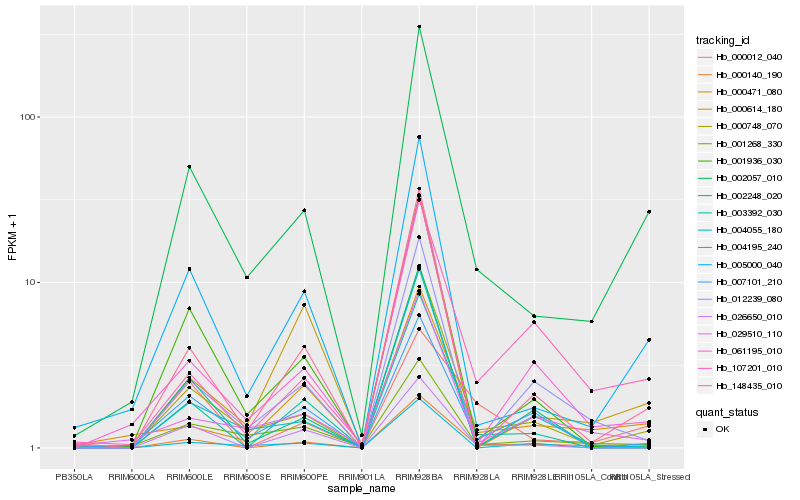
Hb_002057_010 |
0.1427573128 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 4 |
Hb_107201_010 |
0.1630296739 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 5 |
Hb_148435_010 |
0.1784482259 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 6 |
Hb_003392_030 |
0.1827654904 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000471_080 |
0.183335146 |
- |
- |
PREDICTED: probable mitochondrial chaperone BCS1-A [Jatropha curcas] |
| 8 |
Hb_005000_040 |
0.1844834265 |
- |
- |
ACC oxidase ACCO2 [Manihot esculenta] |
| 9 |
Hb_061195_010 |
0.1882140538 |
- |
- |
Galactose oxidase precursor, putative [Ricinus communis] |
| 10 |
Hb_012239_080 |
0.190291764 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase homolog 4-like [Jatropha curcas] |
| 11 |
Hb_029510_110 |
0.2003070407 |
- |
- |
PREDICTED: protein PLANT CADMIUM RESISTANCE 8-like [Jatropha curcas] |
| 12 |
Hb_004195_240 |
0.2067822665 |
- |
- |
Peroxidase 72 precursor, putative [Ricinus communis] |
| 13 |
Hb_007101_210 |
0.2105230311 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 14 |
Hb_001268_330 |
0.215761609 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 15 |
Hb_026650_010 |
0.2168729456 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 16 |
Hb_000012_040 |
0.2181794378 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor RAX2-like [Populus euphratica] |
| 17 |
Hb_001936_030 |
0.2191338475 |
- |
- |
PREDICTED: polygalacturonase At1g48100 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000614_180 |
0.2218134793 |
- |
- |
- |
| 19 |
Hb_004055_180 |
0.2230170278 |
- |
- |
cell wall invertase [Manihot esculenta] |
| 20 |
Hb_002248_020 |
0.2241894295 |
transcription factor |
TF Family: NAC |
PREDICTED: transcription factor JUNGBRUNNEN 1-like isoform X1 [Populus euphratica] |