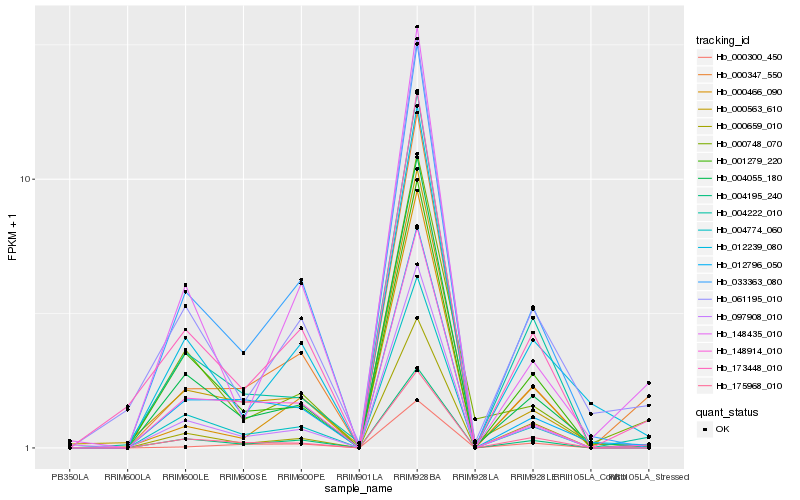
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004195_240 |
0.0 |
- |
- |
Peroxidase 72 precursor, putative [Ricinus communis] |
| 2 |
Hb_000347_550 |
0.0773318153 |
transcription factor |
TF Family: MYB |
Duplicated homeodomain-like superfamily protein, putative [Theobroma cacao] |
| 3 |
Hb_173448_010 |
0.1062969148 |
- |
- |
PREDICTED: galactose oxidase-like [Jatropha curcas] |
| 4 |
Hb_148435_010 |
0.1073905209 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 5 |
Hb_004774_060 |
0.1166052515 |
- |
- |
- |
| 6 |
Hb_061195_010 |
0.1201631292 |
- |
- |
Galactose oxidase precursor, putative [Ricinus communis] |
| 7 |
Hb_148914_010 |
0.1248204999 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 8 |
Hb_033363_080 |
0.1260802926 |
- |
- |
Allene oxide cyclase 3, chloroplast precursor, putative [Ricinus communis] |
| 9 |
Hb_004055_180 |
0.1271669157 |
- |
- |
cell wall invertase [Manihot esculenta] |
| 10 |
Hb_000748_070 |
0.1274060038 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0005s22480g [Populus trichocarpa] |
| 11 |
Hb_000300_450 |
0.1276599996 |
- |
- |
PREDICTED: caffeic acid 3-O-methyltransferase-like isoform X1 [Citrus sinensis] |
| 12 |
Hb_004222_010 |
0.1288051846 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Jatropha curcas] |
| 13 |
Hb_175968_010 |
0.1313607759 |
- |
- |
hypothetical protein JCGZ_21281 [Jatropha curcas] |
| 14 |
Hb_097908_010 |
0.1330676861 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g03230 isoform X2 [Jatropha curcas] |
| 15 |
Hb_012239_080 |
0.1340885252 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase homolog 4-like [Jatropha curcas] |
| 16 |
Hb_012796_050 |
0.1342598021 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
1-deoxyxylulose-5-phosphate synthase, putative [Ricinus communis] |
| 17 |
Hb_001279_220 |
0.140143448 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 18 |
Hb_000659_010 |
0.1422038572 |
- |
- |
PREDICTED: plasma membrane ATPase 2-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_000563_610 |
0.1430014879 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase LIN-1 [Jatropha curcas] |
| 20 |
Hb_000466_090 |
0.1434891984 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RGLG2-like isoform X1 [Jatropha curcas] |