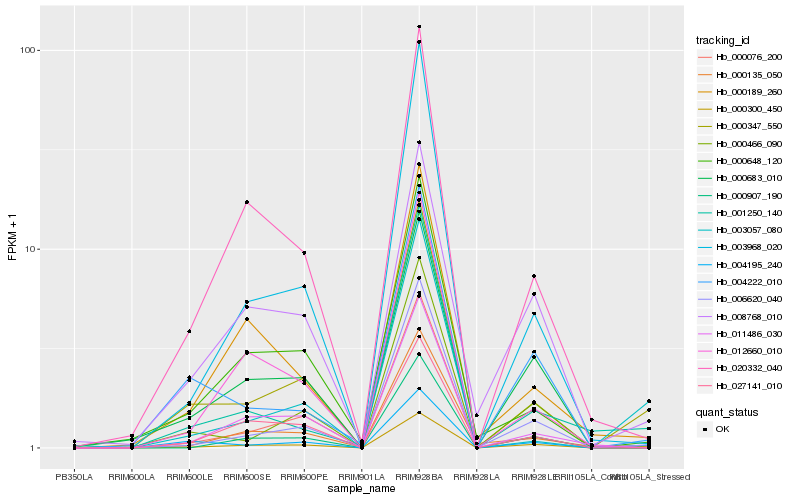
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000300_450 |
0.0 |
- |
- |
PREDICTED: caffeic acid 3-O-methyltransferase-like isoform X1 [Citrus sinensis] |
| 2 |
Hb_000347_550 |
0.0903909506 |
transcription factor |
TF Family: MYB |
Duplicated homeodomain-like superfamily protein, putative [Theobroma cacao] |
| 3 |
Hb_004195_240 |
0.1276599996 |
- |
- |
Peroxidase 72 precursor, putative [Ricinus communis] |
| 4 |
Hb_006620_040 |
0.1280752 |
- |
- |
PREDICTED: probable 6-phosphogluconolactonase 1 [Jatropha curcas] |
| 5 |
Hb_000683_010 |
0.1499801259 |
- |
- |
Interleukin-1 receptor-associated kinase, putative [Ricinus communis] |
| 6 |
Hb_000466_090 |
0.1501962795 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RGLG2-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_000135_050 |
0.1517266015 |
- |
- |
PREDICTED: putative leucine-rich repeat receptor-like protein kinase At2g19210-like isoform X2 [Citrus sinensis] |
| 8 |
Hb_000907_190 |
0.1523397826 |
- |
- |
PREDICTED: uncharacterized protein At4g15970-like [Jatropha curcas] |
| 9 |
Hb_008768_010 |
0.1524330201 |
- |
- |
PREDICTED: equilibrative nucleotide transporter 8 isoform X1 [Jatropha curcas] |
| 10 |
Hb_003968_020 |
0.1550785785 |
- |
- |
PREDICTED: limonoid UDP-glucosyltransferase-like [Glycine max] |
| 11 |
Hb_020332_040 |
0.1560483407 |
- |
- |
PREDICTED: alcohol dehydrogenase 1-like [Jatropha curcas] |
| 12 |
Hb_011486_030 |
0.1630162049 |
- |
- |
branched-chain amino acid aminotransferase, putative [Ricinus communis] |
| 13 |
Hb_000189_260 |
0.1650717778 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000076_200 |
0.1650860287 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_004222_010 |
0.1672495598 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Jatropha curcas] |
| 16 |
Hb_001250_140 |
0.1672975781 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 2 [Jatropha curcas] |
| 17 |
Hb_012660_010 |
0.1678708717 |
- |
- |
hypothetical protein JCGZ_25971 [Jatropha curcas] |
| 18 |
Hb_003057_080 |
0.1687829373 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 1-like [Jatropha curcas] |
| 19 |
Hb_027141_010 |
0.1693968129 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 20 |
Hb_000648_120 |
0.1699513734 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase IV.1 [Vitis vinifera] |