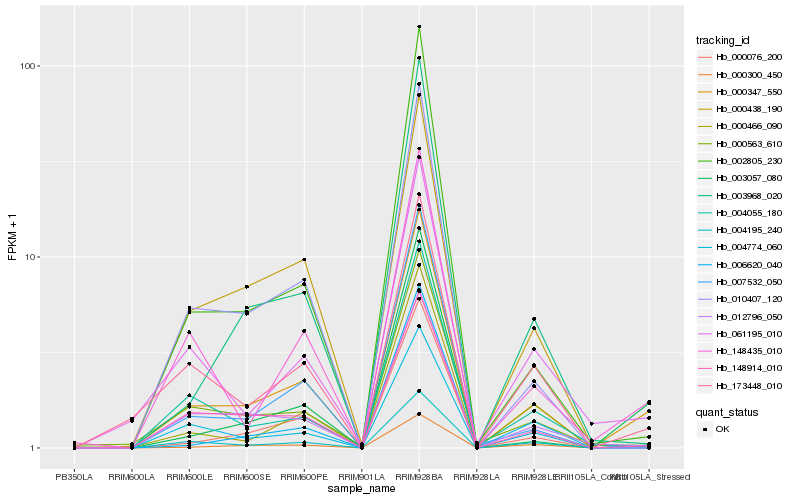
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000347_550 |
0.0 |
transcription factor |
TF Family: MYB |
Duplicated homeodomain-like superfamily protein, putative [Theobroma cacao] |
| 2 |
Hb_004195_240 |
0.0773318153 |
- |
- |
Peroxidase 72 precursor, putative [Ricinus communis] |
| 3 |
Hb_000300_450 |
0.0903909506 |
- |
- |
PREDICTED: caffeic acid 3-O-methyltransferase-like isoform X1 [Citrus sinensis] |
| 4 |
Hb_148435_010 |
0.1238898571 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 5 |
Hb_003057_080 |
0.1301252368 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 1-like [Jatropha curcas] |
| 6 |
Hb_148914_010 |
0.1340377318 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 7 |
Hb_010407_120 |
0.1367276137 |
- |
- |
PREDICTED: glutathione transferase GST 23-like [Jatropha curcas] |
| 8 |
Hb_000466_090 |
0.1390358254 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RGLG2-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_006620_040 |
0.139153807 |
- |
- |
PREDICTED: probable 6-phosphogluconolactonase 1 [Jatropha curcas] |
| 10 |
Hb_173448_010 |
0.1398172098 |
- |
- |
PREDICTED: galactose oxidase-like [Jatropha curcas] |
| 11 |
Hb_000076_200 |
0.1408835674 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000563_610 |
0.1426419527 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase LIN-1 [Jatropha curcas] |
| 13 |
Hb_007532_050 |
0.1438221636 |
- |
- |
9-cis-epoxycarotenoid dioxygenase, putative [Ricinus communis] |
| 14 |
Hb_012796_050 |
0.1459916722 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
1-deoxyxylulose-5-phosphate synthase, putative [Ricinus communis] |
| 15 |
Hb_000438_190 |
0.1460128403 |
- |
- |
PREDICTED: peroxidase 4-like [Jatropha curcas] |
| 16 |
Hb_061195_010 |
0.1469373685 |
- |
- |
Galactose oxidase precursor, putative [Ricinus communis] |
| 17 |
Hb_002805_230 |
0.1487766623 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12 isoform X1 [Jatropha curcas] |
| 18 |
Hb_003968_020 |
0.1491200824 |
- |
- |
PREDICTED: limonoid UDP-glucosyltransferase-like [Glycine max] |
| 19 |
Hb_004774_060 |
0.1500922056 |
- |
- |
- |
| 20 |
Hb_004055_180 |
0.1501353155 |
- |
- |
cell wall invertase [Manihot esculenta] |