| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
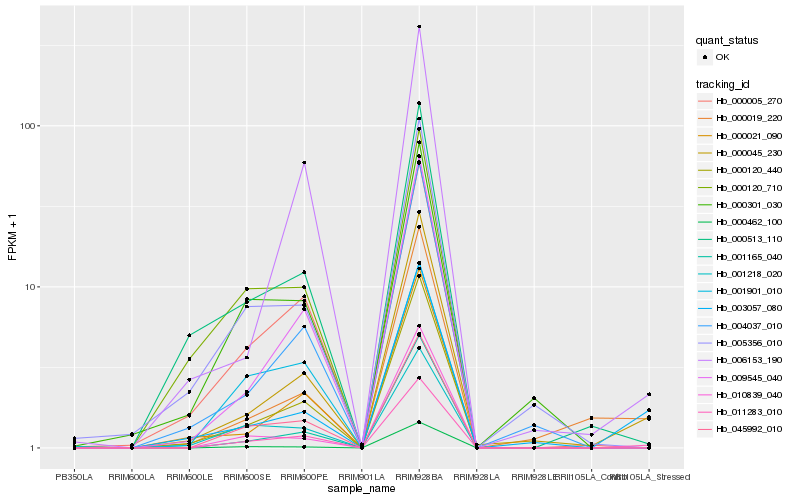
Hb_011283_010 |
0.0 |
- |
- |
PREDICTED: caffeic acid 3-O-methyltransferase-like [Jatropha curcas] |
| 2 |
Hb_045992_010 |
0.1091441491 |
- |
- |
- |
| 3 |
Hb_000045_230 |
0.1098193015 |
- |
- |
PREDICTED: uncharacterized acetyltransferase At3g50280 [Jatropha curcas] |
| 4 |
Hb_000120_440 |
0.1210956659 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000005_270 |
0.1253156204 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase protein 2 precursor, putative [Ricinus communis] |
| 6 |
Hb_009545_040 |
0.1292109032 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001165_040 |
0.1303697574 |
- |
- |
hypothetical protein POPTR_0009s10120g [Populus trichocarpa] |
| 8 |
Hb_003057_080 |
0.1446314664 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 1-like [Jatropha curcas] |
| 9 |
Hb_000021_090 |
0.149160508 |
- |
- |
- |
| 10 |
Hb_004037_010 |
0.1493378991 |
- |
- |
PREDICTED: uncharacterized protein LOC105632369 [Jatropha curcas] |
| 11 |
Hb_000301_030 |
0.1524577385 |
- |
- |
PREDICTED: pectate lyase-like [Jatropha curcas] |
| 12 |
Hb_006153_190 |
0.1535082349 |
- |
- |
PREDICTED: 14 kDa proline-rich protein DC2.15-like [Gossypium raimondii] |
| 13 |
Hb_000513_110 |
0.1535196558 |
- |
- |
PREDICTED: germin-like protein subfamily 1 member 13 [Populus euphratica] |
| 14 |
Hb_000462_100 |
0.1540729608 |
- |
- |
Leucoanthocyanidin dioxygenase, putative [Ricinus communis] |
| 15 |
Hb_000120_710 |
0.154556373 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 16 |
Hb_001901_010 |
0.1546101261 |
- |
- |
PREDICTED: alpha carbonic anhydrase 7-like [Jatropha curcas] |
| 17 |
Hb_001218_020 |
0.1546265309 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0005s17080g [Populus trichocarpa] |
| 18 |
Hb_005356_010 |
0.1552461319 |
- |
- |
hypothetical protein POPTR_0003s16550g [Populus trichocarpa] |
| 19 |
Hb_000019_220 |
0.1562531637 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF113-like [Jatropha curcas] |
| 20 |
Hb_010839_040 |
0.1577677014 |
- |
- |
hypothetical protein JCGZ_20868 [Jatropha curcas] |