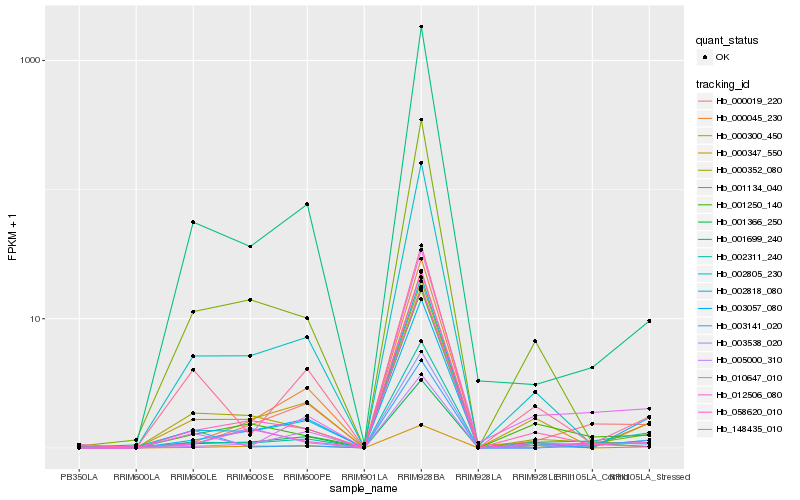
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002311_240 |
0.0 |
- |
- |
hypothetical protein JCGZ_18937 [Jatropha curcas] |
| 2 |
Hb_010647_010 |
0.1004525817 |
- |
- |
PREDICTED: protein NP24-like [Eucalyptus grandis] |
| 3 |
Hb_000019_220 |
0.1160796834 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF113-like [Jatropha curcas] |
| 4 |
Hb_001250_140 |
0.1190749442 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 2 [Jatropha curcas] |
| 5 |
Hb_003057_080 |
0.1234793845 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 1-like [Jatropha curcas] |
| 6 |
Hb_005000_310 |
0.1471615502 |
- |
- |
PREDICTED: uncharacterized protein LOC105637900 [Jatropha curcas] |
| 7 |
Hb_000347_550 |
0.1560904842 |
transcription factor |
TF Family: MYB |
Duplicated homeodomain-like superfamily protein, putative [Theobroma cacao] |
| 8 |
Hb_000045_230 |
0.1593291529 |
- |
- |
PREDICTED: uncharacterized acetyltransferase At3g50280 [Jatropha curcas] |
| 9 |
Hb_000352_080 |
0.167104367 |
- |
- |
PREDICTED: lipid transfer-like protein VAS [Jatropha curcas] |
| 10 |
Hb_058620_010 |
0.1692536944 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 11 |
Hb_002818_080 |
0.1714310777 |
transcription factor |
TF Family: AUX/IAA |
Auxin-responsive protein IAA20, putative [Ricinus communis] |
| 12 |
Hb_001699_240 |
0.1737736509 |
- |
- |
PREDICTED: naringenin,2-oxoglutarate 3-dioxygenase [Jatropha curcas] |
| 13 |
Hb_012506_080 |
0.1771692725 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 14 |
Hb_002805_230 |
0.1807103512 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001366_250 |
0.1812514732 |
- |
- |
PREDICTED: mannan endo-1,4-beta-mannosidase 4-like [Jatropha curcas] |
| 16 |
Hb_003538_020 |
0.1821007609 |
- |
- |
(S)-N-methylcoclaurine 3'-hydroxylase isozyme, putative [Ricinus communis] |
| 17 |
Hb_003141_020 |
0.1833514065 |
- |
- |
PREDICTED: chlorophyllase-1-like [Jatropha curcas] |
| 18 |
Hb_148435_010 |
0.1853625259 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 19 |
Hb_001134_040 |
0.1864513667 |
- |
- |
PREDICTED: EG45-like domain containing protein [Jatropha curcas] |
| 20 |
Hb_000300_450 |
0.1876403997 |
- |
- |
PREDICTED: caffeic acid 3-O-methyltransferase-like isoform X1 [Citrus sinensis] |