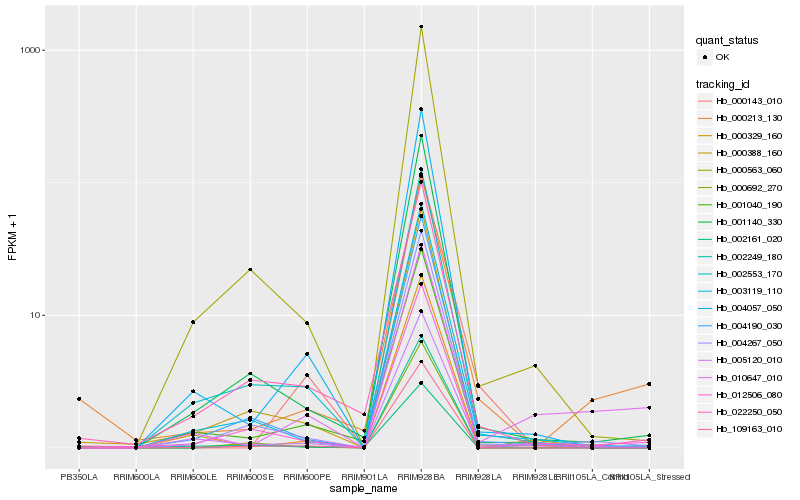
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000213_130 |
0.0 |
transcription factor |
TF Family: HSF |
Heat Stress Transcription Factor family protein [Populus trichocarpa] |
| 2 |
Hb_012506_080 |
0.1457607597 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 3 |
Hb_004267_050 |
0.1493790852 |
- |
- |
hypothetical protein JCGZ_04357 [Jatropha curcas] |
| 4 |
Hb_022250_050 |
0.1505428862 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001140_330 |
0.1509474814 |
- |
- |
PREDICTED: uncharacterized protein LOC105142675 [Populus euphratica] |
| 6 |
Hb_005120_010 |
0.1528373961 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 7 |
Hb_001040_190 |
0.1578175304 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor B-2a-like [Jatropha curcas] |
| 8 |
Hb_003119_110 |
0.1587535992 |
- |
- |
PREDICTED: feruloyl CoA ortho-hydroxylase 2-like [Jatropha curcas] |
| 9 |
Hb_004190_030 |
0.1632505361 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 10 |
Hb_004057_050 |
0.1642810085 |
- |
- |
PREDICTED: cytochrome P450 76A1-like [Jatropha curcas] |
| 11 |
Hb_010647_010 |
0.1643180284 |
- |
- |
PREDICTED: protein NP24-like [Eucalyptus grandis] |
| 12 |
Hb_000143_010 |
0.1651825505 |
- |
- |
hypothetical protein JCGZ_16070 [Jatropha curcas] |
| 13 |
Hb_000388_160 |
0.1681612582 |
- |
- |
xyloglucan endo-1 family protein [Populus trichocarpa] |
| 14 |
Hb_000692_270 |
0.1688647907 |
- |
- |
PREDICTED: ornithine decarboxylase-like [Jatropha curcas] |
| 15 |
Hb_000563_060 |
0.1690060899 |
- |
- |
PREDICTED: uncharacterized protein LOC100306381 isoform X1 [Glycine max] |
| 16 |
Hb_002161_020 |
0.169127472 |
transcription factor |
TF Family: bZIP |
hypothetical protein POPTR_0006s05770g, partial [Populus trichocarpa] |
| 17 |
Hb_002553_170 |
0.1692428567 |
- |
- |
PREDICTED: early nodulin-75-like [Jatropha curcas] |
| 18 |
Hb_109163_010 |
0.1693830645 |
- |
- |
polyphenol oxidase [Hevea brasiliensis] |
| 19 |
Hb_002249_180 |
0.1695595896 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 20 |
Hb_000329_160 |
0.1704174955 |
- |
- |
PREDICTED: pectinesterase 2 isoform X1 [Jatropha curcas] |