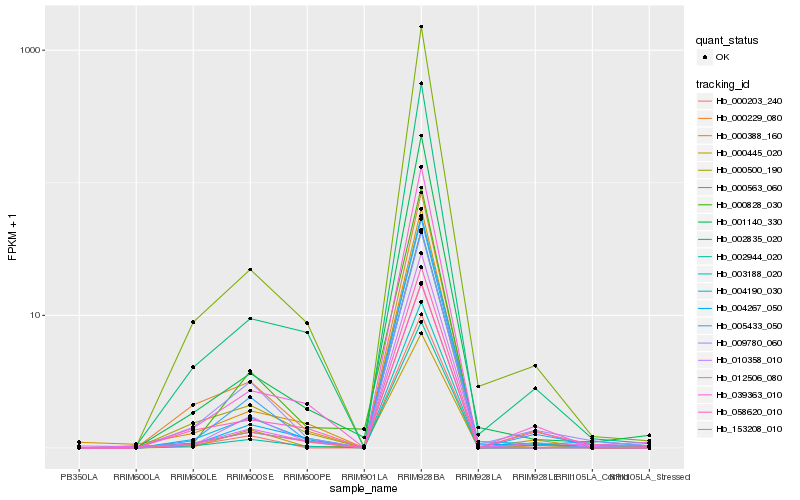
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012506_080 |
0.0 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 2 |
Hb_005433_050 |
0.0721592046 |
- |
- |
PREDICTED: uncharacterized protein At1g66480-like [Populus euphratica] |
| 3 |
Hb_000388_160 |
0.0788836299 |
- |
- |
xyloglucan endo-1 family protein [Populus trichocarpa] |
| 4 |
Hb_000203_240 |
0.0803303787 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 5 |
Hb_002835_020 |
0.0833548977 |
- |
- |
PREDICTED: cytochrome P450 71A1-like [Jatropha curcas] |
| 6 |
Hb_004267_050 |
0.0880247721 |
- |
- |
hypothetical protein JCGZ_04357 [Jatropha curcas] |
| 7 |
Hb_039363_010 |
0.0894631216 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 8 |
Hb_001140_330 |
0.0896180305 |
- |
- |
PREDICTED: uncharacterized protein LOC105142675 [Populus euphratica] |
| 9 |
Hb_000563_060 |
0.0900957202 |
- |
- |
PREDICTED: uncharacterized protein LOC100306381 isoform X1 [Glycine max] |
| 10 |
Hb_002944_020 |
0.0905317022 |
transcription factor |
TF Family: bZIP |
hypothetical protein POPTR_0006s05770g, partial [Populus trichocarpa] |
| 11 |
Hb_000229_080 |
0.0928481479 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 12 |
Hb_058620_010 |
0.0941600499 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 13 |
Hb_000500_190 |
0.0947307094 |
- |
- |
hypothetical protein JCGZ_21655 [Jatropha curcas] |
| 14 |
Hb_009780_060 |
0.0955055766 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranyl geranyl pyrophosphate synthase, putative [Ricinus communis] |
| 15 |
Hb_000828_030 |
0.0964093874 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 16 |
Hb_153208_010 |
0.0968678665 |
- |
- |
Cationic peroxidase 2 precursor, putative [Ricinus communis] |
| 17 |
Hb_004190_030 |
0.0971705042 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_010358_010 |
0.0979274423 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 19 |
Hb_000445_020 |
0.0983163157 |
- |
- |
GABA-specific permease, putative [Ricinus communis] |
| 20 |
Hb_003188_020 |
0.0986243384 |
- |
- |
chitinase, putative [Ricinus communis] |