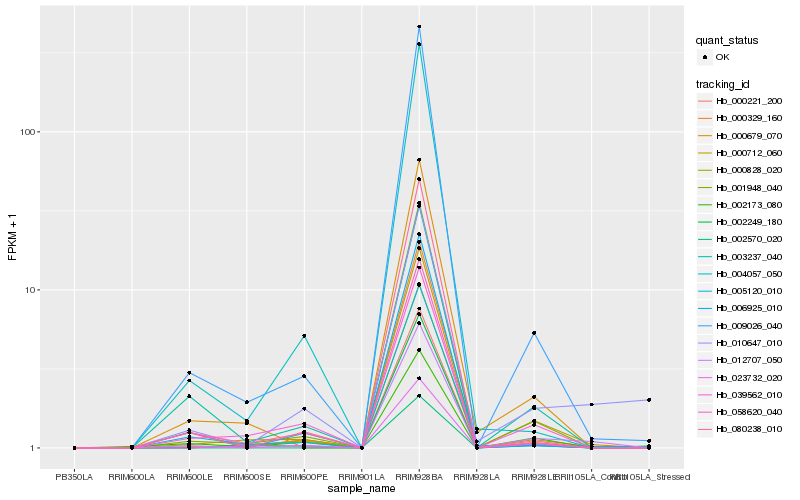
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012707_050 |
0.0 |
- |
- |
hypothetical protein RCOM_1341590 [Ricinus communis] |
| 2 |
Hb_000712_060 |
0.0918683762 |
- |
- |
PREDICTED: uncharacterized protein LOC105631517 [Jatropha curcas] |
| 3 |
Hb_000828_020 |
0.1096133962 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 4 |
Hb_001948_040 |
0.1118649541 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranyl geranyl pyrophosphate synthase, putative [Ricinus communis] |
| 5 |
Hb_009026_040 |
0.1155425891 |
- |
- |
pathogenesis-related family protein [Populus trichocarpa] |
| 6 |
Hb_000221_200 |
0.1220715343 |
- |
- |
hypothetical protein JCGZ_15819 [Jatropha curcas] |
| 7 |
Hb_002570_020 |
0.1264498523 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 8 |
Hb_003237_040 |
0.1320771827 |
- |
- |
Ent-kaurene synthase B, chloroplast precursor, putative [Ricinus communis] |
| 9 |
Hb_080238_010 |
0.1368076745 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 10 |
Hb_000679_070 |
0.1374584957 |
- |
- |
hypothetical protein POPTR_0006s26840g [Populus trichocarpa] |
| 11 |
Hb_010647_010 |
0.1376619176 |
- |
- |
PREDICTED: protein NP24-like [Eucalyptus grandis] |
| 12 |
Hb_005120_010 |
0.1398385762 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 13 |
Hb_006925_010 |
0.1413524665 |
- |
- |
PREDICTED: uncharacterized protein LOC101777526 [Setaria italica] |
| 14 |
Hb_058620_040 |
0.1428001689 |
- |
- |
Myrcene synthase, chloroplastic, putative isoform 1 [Theobroma cacao] |
| 15 |
Hb_000329_160 |
0.1439694597 |
- |
- |
PREDICTED: pectinesterase 2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_023732_020 |
0.1455862796 |
- |
- |
PREDICTED: fatty acyl-CoA reductase 3-like [Jatropha curcas] |
| 17 |
Hb_002249_180 |
0.1461411268 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 18 |
Hb_039562_010 |
0.1476868483 |
- |
- |
PREDICTED: protein SRG1-like [Jatropha curcas] |
| 19 |
Hb_002173_080 |
0.1492143687 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 20 |
Hb_004057_050 |
0.1495311868 |
- |
- |
PREDICTED: cytochrome P450 76A1-like [Jatropha curcas] |