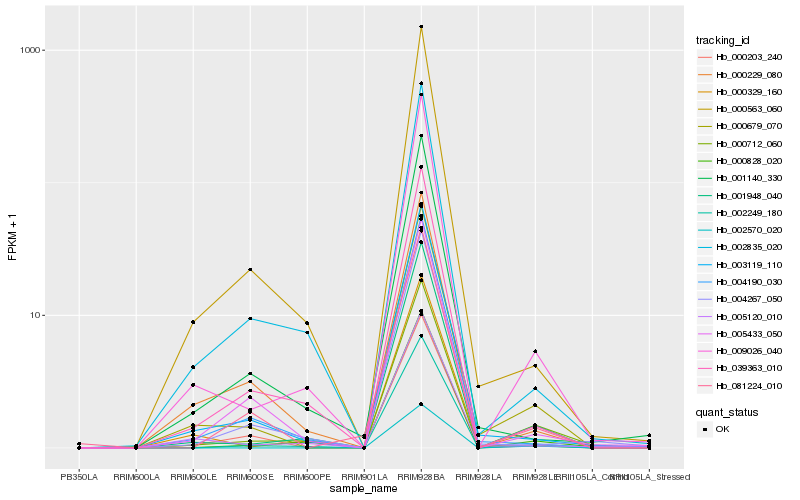
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000679_070 |
0.0 |
- |
- |
hypothetical protein POPTR_0006s26840g [Populus trichocarpa] |
| 2 |
Hb_003119_110 |
0.0770331507 |
- |
- |
PREDICTED: feruloyl CoA ortho-hydroxylase 2-like [Jatropha curcas] |
| 3 |
Hb_009026_040 |
0.0772340008 |
- |
- |
pathogenesis-related family protein [Populus trichocarpa] |
| 4 |
Hb_001948_040 |
0.0783350072 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranyl geranyl pyrophosphate synthase, putative [Ricinus communis] |
| 5 |
Hb_000712_060 |
0.092513909 |
- |
- |
PREDICTED: uncharacterized protein LOC105631517 [Jatropha curcas] |
| 6 |
Hb_004190_030 |
0.0928875704 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 7 |
Hb_000563_060 |
0.0933668516 |
- |
- |
PREDICTED: uncharacterized protein LOC100306381 isoform X1 [Glycine max] |
| 8 |
Hb_002570_020 |
0.0995648593 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 9 |
Hb_000329_160 |
0.1007858669 |
- |
- |
PREDICTED: pectinesterase 2 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002249_180 |
0.1025109832 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 11 |
Hb_000828_020 |
0.1031352594 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 12 |
Hb_005120_010 |
0.1035059459 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 13 |
Hb_039363_010 |
0.1038370877 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 14 |
Hb_001140_330 |
0.1042303187 |
- |
- |
PREDICTED: uncharacterized protein LOC105142675 [Populus euphratica] |
| 15 |
Hb_000203_240 |
0.1061033716 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 16 |
Hb_000229_080 |
0.1072380677 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 17 |
Hb_002835_020 |
0.1078888455 |
- |
- |
PREDICTED: cytochrome P450 71A1-like [Jatropha curcas] |
| 18 |
Hb_005433_050 |
0.1081884254 |
- |
- |
PREDICTED: uncharacterized protein At1g66480-like [Populus euphratica] |
| 19 |
Hb_081224_010 |
0.1092042607 |
- |
- |
hypothetical protein OsJ_17572 [Oryza sativa Japonica Group] |
| 20 |
Hb_004267_050 |
0.1095830468 |
- |
- |
hypothetical protein JCGZ_04357 [Jatropha curcas] |