| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
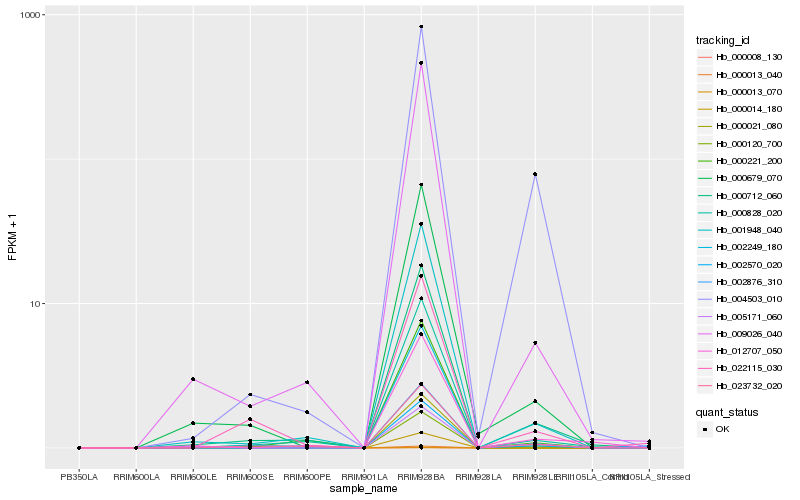
Hb_002876_310 |
0.0 |
- |
- |
hypothetical protein JCGZ_03912 [Jatropha curcas] |
| 2 |
Hb_002570_020 |
0.0952281714 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 3 |
Hb_002249_180 |
0.1185729486 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 4 |
Hb_001948_040 |
0.1328445478 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranyl geranyl pyrophosphate synthase, putative [Ricinus communis] |
| 5 |
Hb_004503_010 |
0.1342066448 |
- |
- |
PREDICTED: EG45-like domain containing protein [Prunus mume] |
| 6 |
Hb_000712_060 |
0.137900256 |
- |
- |
PREDICTED: uncharacterized protein LOC105631517 [Jatropha curcas] |
| 7 |
Hb_009026_040 |
0.1393132709 |
- |
- |
pathogenesis-related family protein [Populus trichocarpa] |
| 8 |
Hb_000679_070 |
0.1417958766 |
- |
- |
hypothetical protein POPTR_0006s26840g [Populus trichocarpa] |
| 9 |
Hb_000828_020 |
0.1477151333 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 10 |
Hb_023732_020 |
0.1542399211 |
- |
- |
PREDICTED: fatty acyl-CoA reductase 3-like [Jatropha curcas] |
| 11 |
Hb_022115_030 |
0.1559309092 |
- |
- |
PREDICTED: umecyanin-like [Jatropha curcas] |
| 12 |
Hb_012707_050 |
0.1578345797 |
- |
- |
hypothetical protein RCOM_1341590 [Ricinus communis] |
| 13 |
Hb_000221_200 |
0.1584007076 |
- |
- |
hypothetical protein JCGZ_15819 [Jatropha curcas] |
| 14 |
Hb_005171_060 |
0.1587987607 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 15 |
Hb_000120_700 |
0.1592594117 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 16 |
Hb_000008_130 |
0.1648040212 |
- |
- |
PREDICTED: uncharacterized protein LOC105630320 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000013_040 |
0.1648040212 |
- |
- |
- |
| 18 |
Hb_000013_070 |
0.1648040212 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 19 |
Hb_000014_180 |
0.1648040212 |
- |
- |
PREDICTED: endo-1,4-beta-xylanase D-like [Fragaria vesca subsp. vesca] |
| 20 |
Hb_000021_080 |
0.1648040212 |
- |
- |
- |