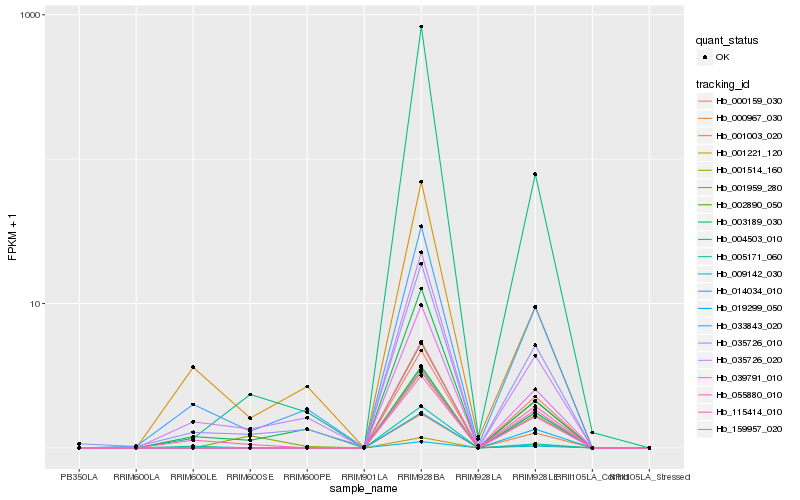
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_039791_010 |
0.0 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_001221_120 |
0.0725157275 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 3 |
Hb_002890_050 |
0.0749682034 |
- |
- |
- |
| 4 |
Hb_014034_010 |
0.0759353466 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 5 |
Hb_001514_160 |
0.0806147263 |
- |
- |
hypothetical protein JCGZ_16070 [Jatropha curcas] |
| 6 |
Hb_009142_030 |
0.0825500149 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 7 |
Hb_055880_010 |
0.0855421901 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 8 |
Hb_004503_010 |
0.0938231176 |
- |
- |
PREDICTED: EG45-like domain containing protein [Prunus mume] |
| 9 |
Hb_159957_020 |
0.1088129003 |
- |
- |
- |
| 10 |
Hb_000159_030 |
0.1105084587 |
- |
- |
putative glycosyltransferase, partial [Aralia elata] |
| 11 |
Hb_000967_030 |
0.128013994 |
- |
- |
- |
| 12 |
Hb_115414_010 |
0.1335115216 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Jatropha curcas] |
| 13 |
Hb_035726_010 |
0.1529901262 |
- |
- |
Insulin-degrading enzyme, putative [Ricinus communis] |
| 14 |
Hb_001959_280 |
0.1575061797 |
- |
- |
PREDICTED: uncharacterized protein LOC105638432 [Jatropha curcas] |
| 15 |
Hb_005171_060 |
0.1653242932 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 16 |
Hb_019299_050 |
0.1673690525 |
- |
- |
- |
| 17 |
Hb_035726_020 |
0.1704659509 |
- |
- |
Insulinase (Peptidase family M16) family protein [Theobroma cacao] |
| 18 |
Hb_033843_020 |
0.1705461813 |
- |
- |
lysosomal alpha-mannosidase, putative [Ricinus communis] |
| 19 |
Hb_003189_030 |
0.1794434172 |
- |
- |
hypothetical protein Csa_4G304750 [Cucumis sativus] |
| 20 |
Hb_001003_020 |
0.1839007133 |
- |
- |
PREDICTED: metalloendoproteinase 1-like [Populus euphratica] |