| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
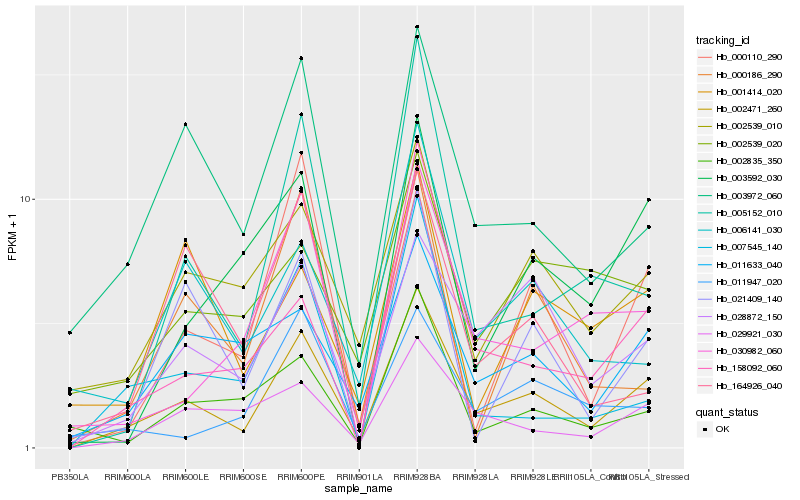
Hb_002471_260 |
0.0 |
transcription factor |
TF Family: PLATZ |
PREDICTED: uncharacterized protein LOC105634154 [Jatropha curcas] |
| 2 |
Hb_028872_150 |
0.1773699834 |
- |
- |
PREDICTED: uncharacterized protein LOC105137493 [Populus euphratica] |
| 3 |
Hb_029921_030 |
0.1783731769 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_003972_060 |
0.1787073742 |
- |
- |
PREDICTED: UDP-arabinopyranose mutase 3 [Populus euphratica] |
| 5 |
Hb_002539_010 |
0.1849058387 |
- |
- |
hypothetical protein JCGZ_15711 [Jatropha curcas] |
| 6 |
Hb_011633_040 |
0.1874246996 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase SD1-8 [Jatropha curcas] |
| 7 |
Hb_021409_140 |
0.2007206054 |
- |
- |
PREDICTED: trehalase isoform X1 [Jatropha curcas] |
| 8 |
Hb_005152_010 |
0.2029854754 |
- |
- |
hypothetical protein JCGZ_22048 [Jatropha curcas] |
| 9 |
Hb_158092_060 |
0.204374236 |
- |
- |
aspartate semialdehyde dehydrogenase, putative [Ricinus communis] |
| 10 |
Hb_030982_060 |
0.2093117999 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_006141_030 |
0.2121125157 |
- |
- |
- |
| 12 |
Hb_011947_020 |
0.2142222066 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 13 |
Hb_002835_350 |
0.2165466099 |
- |
- |
PREDICTED: uncharacterized protein LOC105640319 [Jatropha curcas] |
| 14 |
Hb_003592_030 |
0.2167202161 |
- |
- |
PREDICTED: dCTP pyrophosphatase 1-like [Jatropha curcas] |
| 15 |
Hb_000110_290 |
0.2190177413 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_007545_140 |
0.219653574 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105141503 [Populus euphratica] |
| 17 |
Hb_002539_020 |
0.2243924138 |
- |
- |
hypothetical protein CISIN_1g007216mg [Citrus sinensis] |
| 18 |
Hb_000186_290 |
0.2246469076 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 8 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001414_020 |
0.2269246872 |
- |
- |
PREDICTED: CRAL-TRIO domain-containing protein YKL091C-like isoform X2 [Jatropha curcas] |
| 20 |
Hb_164926_040 |
0.2277650738 |
- |
- |
Beta-1,3-galactosyltransferase sqv-2, putative [Ricinus communis] |