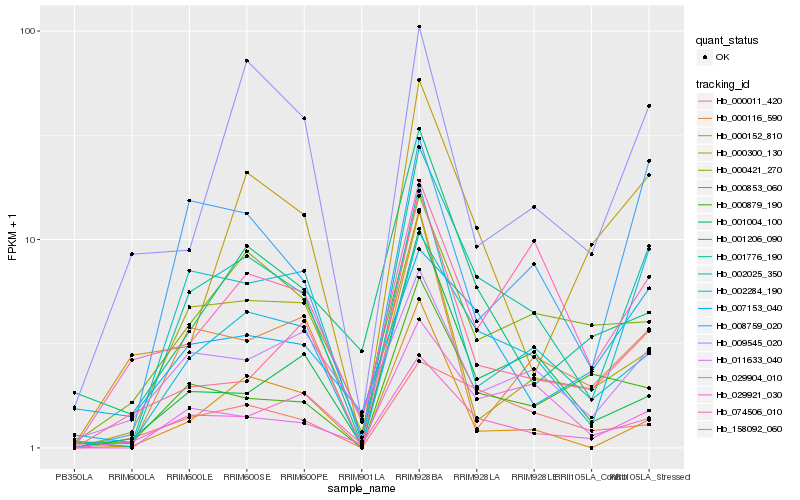
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001776_190 |
0.0 |
- |
- |
PREDICTED: protein kinase 2A, chloroplastic-like [Jatropha curcas] |
| 2 |
Hb_002284_190 |
0.1575503291 |
- |
- |
PREDICTED: glutamate receptor 3.3 [Jatropha curcas] |
| 3 |
Hb_029921_030 |
0.1873909122 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_008759_020 |
0.2033091601 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 2-like [Jatropha curcas] |
| 5 |
Hb_029904_010 |
0.2062990095 |
- |
- |
PREDICTED: uncharacterized protein LOC105635075 [Jatropha curcas] |
| 6 |
Hb_000116_590 |
0.2131694364 |
- |
- |
PREDICTED: splicing factor, suppressor of white-apricot homolog [Malus domestica] |
| 7 |
Hb_011633_040 |
0.2152809552 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase SD1-8 [Jatropha curcas] |
| 8 |
Hb_000853_060 |
0.2179990753 |
- |
- |
PREDICTED: myelin transcription factor 1-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_158092_060 |
0.2236397385 |
- |
- |
aspartate semialdehyde dehydrogenase, putative [Ricinus communis] |
| 10 |
Hb_000011_420 |
0.2248292666 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g29660-like [Citrus sinensis] |
| 11 |
Hb_000879_190 |
0.2249680575 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001004_100 |
0.2266171442 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 13 |
Hb_000300_130 |
0.2276419238 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105141503 [Populus euphratica] |
| 14 |
Hb_007153_040 |
0.2292768678 |
- |
- |
PREDICTED: uncharacterized protein LOC105633515 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000152_810 |
0.229616799 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 16 |
Hb_002025_350 |
0.2302939822 |
- |
- |
PREDICTED: glutamate decarboxylase 4 [Jatropha curcas] |
| 17 |
Hb_001206_090 |
0.2328823977 |
- |
- |
pyruvate decarboxylase [Hevea brasiliensis] |
| 18 |
Hb_074506_010 |
0.2338027335 |
- |
- |
JHL25H03.10 [Jatropha curcas] |
| 19 |
Hb_000421_270 |
0.2383237391 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein 1 [Theobroma cacao] |
| 20 |
Hb_009545_020 |
0.2429125636 |
- |
- |
PREDICTED: uncharacterized protein LOC105632008 [Jatropha curcas] |