| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
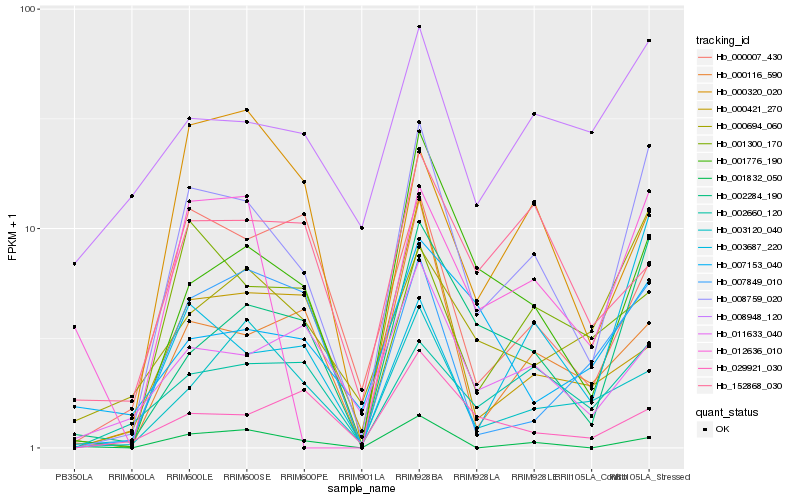
Hb_008759_020 |
0.0 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 2-like [Jatropha curcas] |
| 2 |
Hb_002284_190 |
0.1834565244 |
- |
- |
PREDICTED: glutamate receptor 3.3 [Jatropha curcas] |
| 3 |
Hb_001776_190 |
0.2033091601 |
- |
- |
PREDICTED: protein kinase 2A, chloroplastic-like [Jatropha curcas] |
| 4 |
Hb_002660_120 |
0.2253201722 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase [Populus euphratica] |
| 5 |
Hb_000694_060 |
0.2350306888 |
- |
- |
Rotenone-insensitive NADH-ubiquinone oxidoreductase, mitochondrial precursor, putative [Ricinus communis] |
| 6 |
Hb_003687_220 |
0.238027227 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0008s16660g [Populus trichocarpa] |
| 7 |
Hb_003120_040 |
0.2392707232 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At5g57670 [Jatropha curcas] |
| 8 |
Hb_000116_590 |
0.243238824 |
- |
- |
PREDICTED: splicing factor, suppressor of white-apricot homolog [Malus domestica] |
| 9 |
Hb_011633_040 |
0.2433010369 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase SD1-8 [Jatropha curcas] |
| 10 |
Hb_001300_170 |
0.2447472767 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001832_050 |
0.2456085235 |
- |
- |
PREDICTED: cytokinin dehydrogenase 3 [Jatropha curcas] |
| 12 |
Hb_007849_010 |
0.2514029978 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Populus euphratica] |
| 13 |
Hb_000007_430 |
0.252535384 |
- |
- |
hypothetical protein POPTR_0010s14280g [Populus trichocarpa] |
| 14 |
Hb_012636_010 |
0.2531457809 |
- |
- |
- |
| 15 |
Hb_008948_120 |
0.2566951142 |
- |
- |
PREDICTED: heme oxygenase 1, chloroplastic-like [Jatropha curcas] |
| 16 |
Hb_152868_030 |
0.2646569565 |
- |
- |
abhydrolase domain containing, putative [Ricinus communis] |
| 17 |
Hb_000421_270 |
0.2661650701 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein 1 [Theobroma cacao] |
| 18 |
Hb_029921_030 |
0.268310146 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000320_020 |
0.271709983 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 31 [Jatropha curcas] |
| 20 |
Hb_007153_040 |
0.2759789624 |
- |
- |
PREDICTED: uncharacterized protein LOC105633515 isoform X1 [Jatropha curcas] |