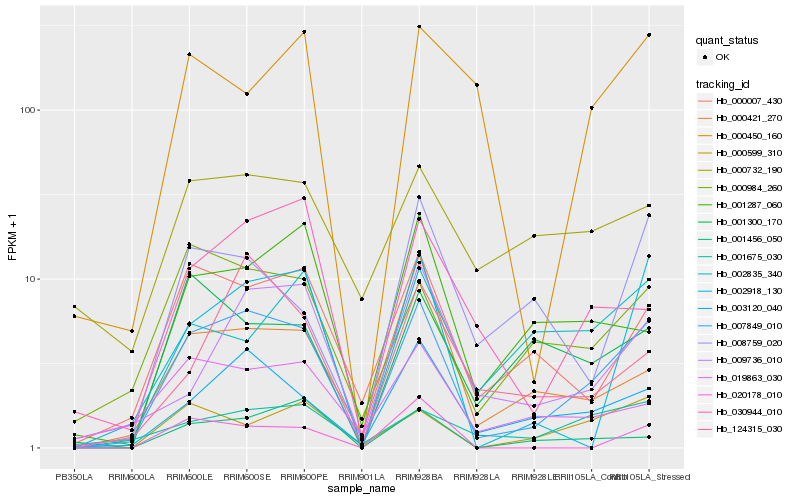
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007849_010 |
0.0 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Populus euphratica] |
| 2 |
Hb_003120_040 |
0.1873404374 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At5g57670 [Jatropha curcas] |
| 3 |
Hb_000007_430 |
0.2056814243 |
- |
- |
hypothetical protein POPTR_0010s14280g [Populus trichocarpa] |
| 4 |
Hb_000984_260 |
0.2135991209 |
- |
- |
ergosterol biosynthetic protein 28 [Jatropha curcas] |
| 5 |
Hb_002918_130 |
0.2145152667 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor 12 [Jatropha curcas] |
| 6 |
Hb_001675_030 |
0.2252473659 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000599_310 |
0.2257414259 |
- |
- |
PREDICTED: pleckstrin homology domain-containing family A member 8-like [Jatropha curcas] |
| 8 |
Hb_009736_010 |
0.2285004239 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_019863_030 |
0.2337400864 |
- |
- |
hypothetical protein JCGZ_00393 [Jatropha curcas] |
| 10 |
Hb_000421_270 |
0.2364447922 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein 1 [Theobroma cacao] |
| 11 |
Hb_030944_010 |
0.2396425489 |
- |
- |
PREDICTED: uncharacterized protein LOC105633455 [Jatropha curcas] |
| 12 |
Hb_001456_050 |
0.2411020723 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 13 |
Hb_001287_060 |
0.2466137846 |
transcription factor |
TF Family: Orphans |
Salt-tolerance protein, putative [Ricinus communis] |
| 14 |
Hb_020178_010 |
0.2487317163 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-related protein A-like [Jatropha curcas] |
| 15 |
Hb_002835_340 |
0.2492197625 |
- |
- |
PREDICTED: peroxisomal primary amine oxidase [Jatropha curcas] |
| 16 |
Hb_001300_170 |
0.2495245983 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_124315_030 |
0.2502350646 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein 21/22, putative [Ricinus communis] |
| 18 |
Hb_008759_020 |
0.2514029978 |
- |
- |
PREDICTED: anthocyanidin 3-O-glucosyltransferase 2-like [Jatropha curcas] |
| 19 |
Hb_000732_190 |
0.2529136186 |
- |
- |
structural molecule, putative [Ricinus communis] |
| 20 |
Hb_000450_160 |
0.2532821581 |
- |
- |
hypothetical protein JCGZ_16746 [Jatropha curcas] |