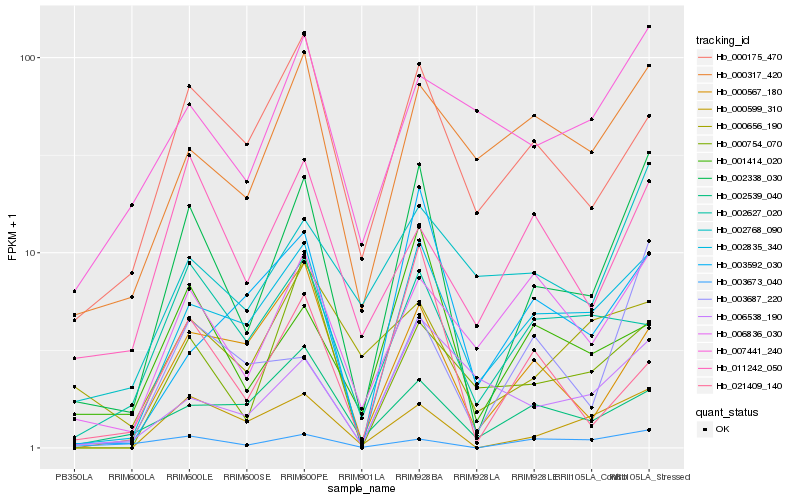
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002338_030 |
0.0 |
- |
- |
PREDICTED: 60S acidic ribosomal protein P1-like [Tarenaya hassleriana] |
| 2 |
Hb_002835_340 |
0.1919476173 |
- |
- |
PREDICTED: peroxisomal primary amine oxidase [Jatropha curcas] |
| 3 |
Hb_000599_310 |
0.1992812091 |
- |
- |
PREDICTED: pleckstrin homology domain-containing family A member 8-like [Jatropha curcas] |
| 4 |
Hb_003673_040 |
0.2393717269 |
- |
- |
dynamin family protein [Populus trichocarpa] |
| 5 |
Hb_006836_030 |
0.2398257613 |
- |
- |
PREDICTED: OTU domain-containing protein DDB_G0284757 [Jatropha curcas] |
| 6 |
Hb_003687_220 |
0.2454191109 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0008s16660g [Populus trichocarpa] |
| 7 |
Hb_000317_420 |
0.2479809849 |
- |
- |
PREDICTED: extradiol ring-cleavage dioxygenase [Jatropha curcas] |
| 8 |
Hb_002768_090 |
0.2495037809 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 11 [Jatropha curcas] |
| 9 |
Hb_000567_180 |
0.252381586 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF1.5 [Jatropha curcas] |
| 10 |
Hb_000175_470 |
0.254906418 |
- |
- |
lactoylglutathione lyase, putative [Ricinus communis] |
| 11 |
Hb_001414_020 |
0.2556076539 |
- |
- |
PREDICTED: CRAL-TRIO domain-containing protein YKL091C-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_002539_040 |
0.262296851 |
- |
- |
SPFH domain-containing protein 2 precursor, putative [Ricinus communis] |
| 13 |
Hb_007441_240 |
0.2642586812 |
- |
- |
ethphon-induced protein [Hevea brasiliensis] |
| 14 |
Hb_011242_050 |
0.2646281674 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2-23 kDa [Jatropha curcas] |
| 15 |
Hb_000754_070 |
0.2659890739 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_006538_190 |
0.266201805 |
- |
- |
PREDICTED: homocysteine S-methyltransferase 1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_003592_030 |
0.2707788083 |
- |
- |
PREDICTED: dCTP pyrophosphatase 1-like [Jatropha curcas] |
| 18 |
Hb_002627_020 |
0.2729796331 |
- |
- |
PREDICTED: TBC1 domain family member 2B [Jatropha curcas] |
| 19 |
Hb_021409_140 |
0.273541476 |
- |
- |
PREDICTED: trehalase isoform X1 [Jatropha curcas] |
| 20 |
Hb_000656_190 |
0.274758583 |
- |
- |
PREDICTED: probable nucleoside diphosphate kinase 5 [Jatropha curcas] |