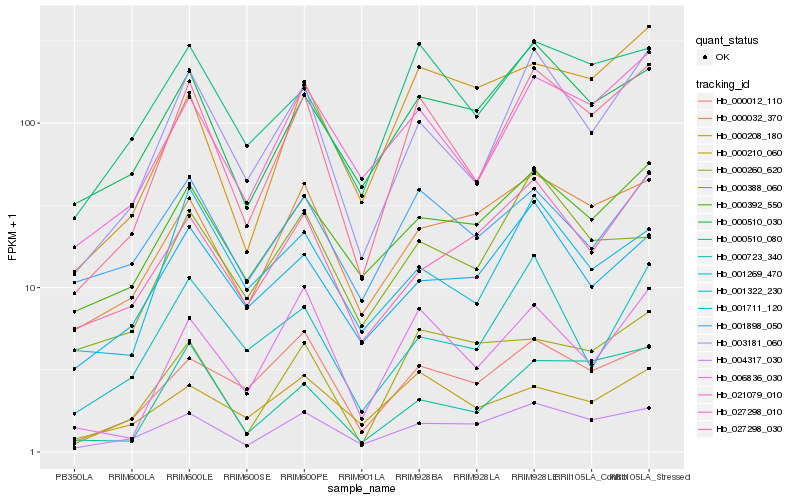
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027298_030 |
0.0 |
- |
- |
PREDICTED: histone H1-like [Jatropha curcas] |
| 2 |
Hb_003181_060 |
0.0967973619 |
- |
- |
PREDICTED: triosephosphate isomerase, cytosolic [Jatropha curcas] |
| 3 |
Hb_021079_010 |
0.1149244551 |
- |
- |
PREDICTED: histone H2B-like [Populus euphratica] |
| 4 |
Hb_004317_030 |
0.130974789 |
- |
- |
PREDICTED: protein CHROMOSOME TRANSMISSION FIDELITY 7 [Jatropha curcas] |
| 5 |
Hb_000510_080 |
0.1328041877 |
- |
- |
hypothetical protein CICLE_v10021453mg [Citrus clementina] |
| 6 |
Hb_006836_030 |
0.1356569068 |
- |
- |
PREDICTED: OTU domain-containing protein DDB_G0284757 [Jatropha curcas] |
| 7 |
Hb_000723_340 |
0.1415338338 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG3 [Jatropha curcas] |
| 8 |
Hb_000510_030 |
0.1424244398 |
- |
- |
Winged-helix DNA-binding transcription factor family protein [Theobroma cacao] |
| 9 |
Hb_000392_550 |
0.1437347573 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 6, chloroplastic isoform X1 [Jatropha curcas] |
| 10 |
Hb_000032_370 |
0.1494877706 |
- |
- |
PREDICTED: nucleoside diphosphate kinase III, chloroplastic/mitochondrial-like [Citrus sinensis] |
| 11 |
Hb_000012_110 |
0.151395476 |
- |
- |
hypothetical protein CICLE_v10008613mg [Citrus clementina] |
| 12 |
Hb_000208_180 |
0.1563658863 |
- |
- |
PREDICTED: histone H2B-like [Populus euphratica] |
| 13 |
Hb_000260_620 |
0.1597714314 |
- |
- |
Thermosensitive gluconokinase, putative [Ricinus communis] |
| 14 |
Hb_001898_050 |
0.1637177278 |
- |
- |
PREDICTED: 60S ribosomal protein L3 [Eucalyptus grandis] |
| 15 |
Hb_000210_060 |
0.1673506899 |
transcription factor |
TF Family: E2F-DP |
hypothetical protein JCGZ_08780 [Jatropha curcas] |
| 16 |
Hb_000388_060 |
0.1680315859 |
- |
- |
fructokinase [Manihot esculenta] |
| 17 |
Hb_001322_230 |
0.1699147447 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 4, chloroplastic [Populus euphratica] |
| 18 |
Hb_001711_120 |
0.1705556464 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 4, chloroplastic [Jatropha curcas] |
| 19 |
Hb_001269_470 |
0.1706511376 |
- |
- |
PREDICTED: uncharacterized protein LOC105630313 [Jatropha curcas] |
| 20 |
Hb_027298_010 |
0.1724362757 |
- |
- |
conserved hypothetical protein [Ricinus communis] |