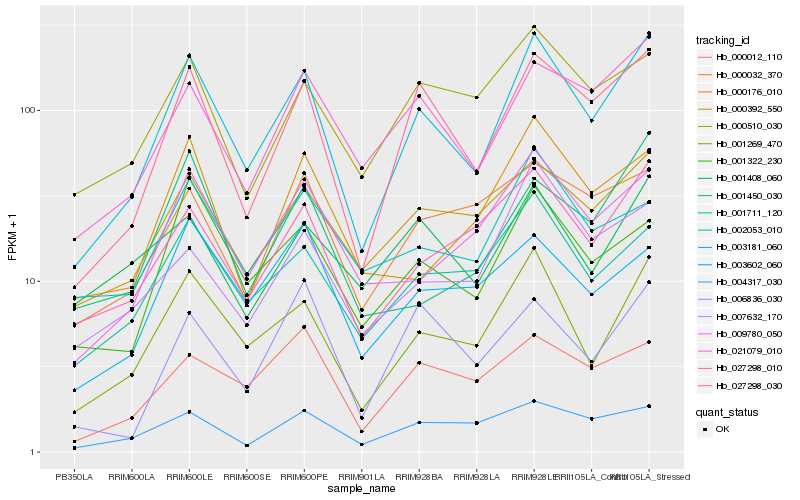
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003181_060 |
0.0 |
- |
- |
PREDICTED: triosephosphate isomerase, cytosolic [Jatropha curcas] |
| 2 |
Hb_001269_470 |
0.0937209311 |
- |
- |
PREDICTED: uncharacterized protein LOC105630313 [Jatropha curcas] |
| 3 |
Hb_027298_030 |
0.0967973619 |
- |
- |
PREDICTED: histone H1-like [Jatropha curcas] |
| 4 |
Hb_021079_010 |
0.1296195142 |
- |
- |
PREDICTED: histone H2B-like [Populus euphratica] |
| 5 |
Hb_027298_010 |
0.1343303961 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001711_120 |
0.13570507 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 4, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000392_550 |
0.1382775395 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 6, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_009780_050 |
0.1393073337 |
- |
- |
PREDICTED: glutathione S-transferase DHAR3, chloroplastic [Jatropha curcas] |
| 9 |
Hb_001322_230 |
0.1398505704 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 4, chloroplastic [Populus euphratica] |
| 10 |
Hb_004317_030 |
0.1539217274 |
- |
- |
PREDICTED: protein CHROMOSOME TRANSMISSION FIDELITY 7 [Jatropha curcas] |
| 11 |
Hb_000510_030 |
0.1547355034 |
- |
- |
Winged-helix DNA-binding transcription factor family protein [Theobroma cacao] |
| 12 |
Hb_000012_110 |
0.1576472357 |
- |
- |
hypothetical protein CICLE_v10008613mg [Citrus clementina] |
| 13 |
Hb_001450_030 |
0.1589593868 |
- |
- |
PREDICTED: uncharacterized protein LOC105650827 [Jatropha curcas] |
| 14 |
Hb_000176_010 |
0.1602364416 |
- |
- |
PREDICTED: 50S ribosomal protein L35, chloroplastic [Jatropha curcas] |
| 15 |
Hb_003602_060 |
0.1624676093 |
- |
- |
PREDICTED: phosphoglycerate mutase-like protein 1 [Jatropha curcas] |
| 16 |
Hb_006836_030 |
0.1635115978 |
- |
- |
PREDICTED: OTU domain-containing protein DDB_G0284757 [Jatropha curcas] |
| 17 |
Hb_001408_060 |
0.1640844622 |
- |
- |
PREDICTED: 60S ribosomal protein L3 [Eucalyptus grandis] |
| 18 |
Hb_002053_010 |
0.1641600843 |
- |
- |
PREDICTED: clp protease-related protein At4g12060, chloroplastic-like [Jatropha curcas] |
| 19 |
Hb_000032_370 |
0.1641931818 |
- |
- |
PREDICTED: nucleoside diphosphate kinase III, chloroplastic/mitochondrial-like [Citrus sinensis] |
| 20 |
Hb_007632_170 |
0.1643795124 |
- |
- |
hypothetical protein JCGZ_01028 [Jatropha curcas] |