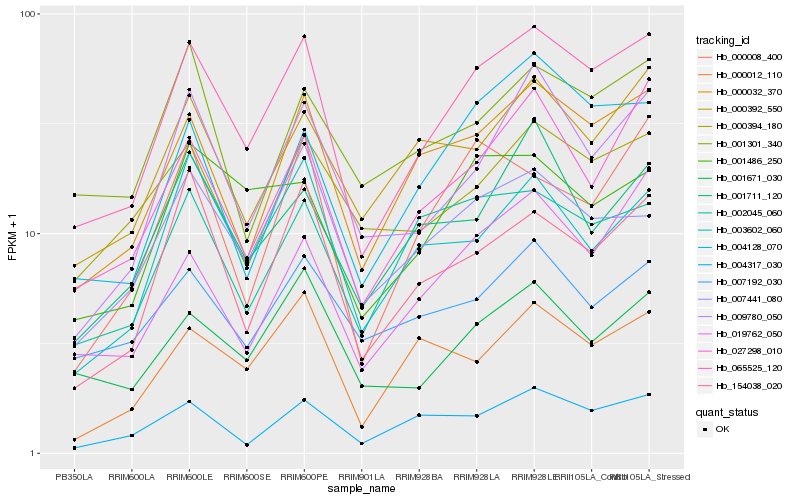
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_065525_120 |
0.0 |
- |
- |
dtdp-glucose 4-6-dehydratase, putative [Ricinus communis] |
| 2 |
Hb_000032_370 |
0.090088156 |
- |
- |
PREDICTED: nucleoside diphosphate kinase III, chloroplastic/mitochondrial-like [Citrus sinensis] |
| 3 |
Hb_027298_010 |
0.1139545411 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_009780_050 |
0.1141727131 |
- |
- |
PREDICTED: glutathione S-transferase DHAR3, chloroplastic [Jatropha curcas] |
| 5 |
Hb_154038_020 |
0.1183923267 |
- |
- |
hypothetical protein JCGZ_05648 [Jatropha curcas] |
| 6 |
Hb_001671_030 |
0.1207745803 |
- |
- |
PREDICTED: calcium-transporting ATPase, endoplasmic reticulum-type [Jatropha curcas] |
| 7 |
Hb_007441_080 |
0.1226547173 |
- |
- |
PREDICTED: ELMO domain-containing protein C-like [Jatropha curcas] |
| 8 |
Hb_004317_030 |
0.1230835356 |
- |
- |
PREDICTED: protein CHROMOSOME TRANSMISSION FIDELITY 7 [Jatropha curcas] |
| 9 |
Hb_002045_060 |
0.1258046367 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] flavoprotein 2, mitochondrial [Jatropha curcas] |
| 10 |
Hb_003602_060 |
0.1258995751 |
- |
- |
PREDICTED: phosphoglycerate mutase-like protein 1 [Jatropha curcas] |
| 11 |
Hb_000392_550 |
0.1265685321 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 6, chloroplastic isoform X1 [Jatropha curcas] |
| 12 |
Hb_004128_070 |
0.1284507155 |
- |
- |
PREDICTED: thioredoxin-like protein AAED1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_001486_250 |
0.1290703068 |
- |
- |
PREDICTED: uncharacterized protein LOC105632605 [Jatropha curcas] |
| 14 |
Hb_000394_180 |
0.1291422401 |
- |
- |
PREDICTED: uncharacterized protein LOC105636995 [Jatropha curcas] |
| 15 |
Hb_019762_050 |
0.1325071462 |
transcription factor |
TF Family: CSD |
PREDICTED: glycine-rich protein 2-like [Jatropha curcas] |
| 16 |
Hb_001711_120 |
0.1341904886 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 4, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000012_110 |
0.1347439218 |
- |
- |
hypothetical protein CICLE_v10008613mg [Citrus clementina] |
| 18 |
Hb_007192_030 |
0.1348882951 |
- |
- |
PREDICTED: manganese-dependent ADP-ribose/CDP-alcohol diphosphatase [Jatropha curcas] |
| 19 |
Hb_001301_340 |
0.1356896078 |
- |
- |
endoribonuclease L-PSP family protein [Populus trichocarpa] |
| 20 |
Hb_000008_400 |
0.1364243184 |
- |
- |
PREDICTED: classical arabinogalactan protein 10-like [Nicotiana tomentosiformis] |