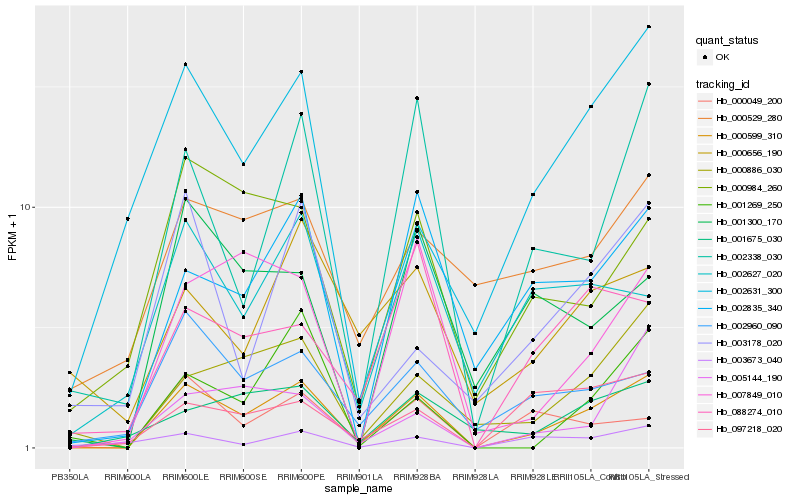
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000599_310 |
0.0 |
- |
- |
PREDICTED: pleckstrin homology domain-containing family A member 8-like [Jatropha curcas] |
| 2 |
Hb_002338_030 |
0.1992812091 |
- |
- |
PREDICTED: 60S acidic ribosomal protein P1-like [Tarenaya hassleriana] |
| 3 |
Hb_002835_340 |
0.2110070632 |
- |
- |
PREDICTED: peroxisomal primary amine oxidase [Jatropha curcas] |
| 4 |
Hb_000886_030 |
0.2116236582 |
- |
- |
PREDICTED: uncharacterized protein LOC105113410 [Populus euphratica] |
| 5 |
Hb_007849_010 |
0.2257414259 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Populus euphratica] |
| 6 |
Hb_000049_200 |
0.2280072319 |
- |
- |
PREDICTED: uncharacterized protein LOC105644462 [Jatropha curcas] |
| 7 |
Hb_002960_090 |
0.2309271064 |
- |
- |
protein with unknown function [Ricinus communis] |
| 8 |
Hb_001269_250 |
0.2320283796 |
- |
- |
PREDICTED: factor of DNA methylation 1-like [Jatropha curcas] |
| 9 |
Hb_001675_030 |
0.2348067195 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000984_260 |
0.2364268912 |
- |
- |
ergosterol biosynthetic protein 28 [Jatropha curcas] |
| 11 |
Hb_002627_020 |
0.2389273477 |
- |
- |
PREDICTED: TBC1 domain family member 2B [Jatropha curcas] |
| 12 |
Hb_088274_010 |
0.2418428693 |
- |
- |
PREDICTED: transcription factor BEE 1-like [Jatropha curcas] |
| 13 |
Hb_002631_300 |
0.2438782738 |
- |
- |
PREDICTED: uncharacterized protein LOC105646181 [Jatropha curcas] |
| 14 |
Hb_097218_020 |
0.2467121961 |
- |
- |
PREDICTED: replication protein A 70 kDa DNA-binding subunit E [Jatropha curcas] |
| 15 |
Hb_005144_190 |
0.2526476061 |
- |
- |
hypothetical protein F383_09558 [Gossypium arboreum] |
| 16 |
Hb_001300_170 |
0.2533984456 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003178_020 |
0.2536128194 |
transcription factor |
TF Family: B3 |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000529_280 |
0.2554045991 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_003673_040 |
0.2564796447 |
- |
- |
dynamin family protein [Populus trichocarpa] |
| 20 |
Hb_000656_190 |
0.2578346551 |
- |
- |
PREDICTED: probable nucleoside diphosphate kinase 5 [Jatropha curcas] |