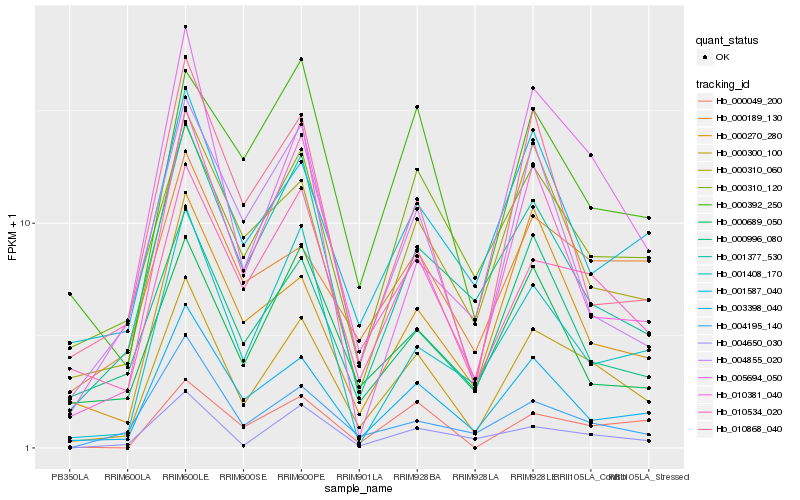
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000300_100 |
0.0 |
- |
- |
NADH-cytochrome b5 reductase 1 [Morus notabilis] |
| 2 |
Hb_010534_020 |
0.1250923568 |
- |
- |
PREDICTED: protein XRI1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_003398_040 |
0.1329450108 |
- |
- |
PREDICTED: actin-related protein 5 isoform X1 [Jatropha curcas] |
| 4 |
Hb_010381_040 |
0.1528467873 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 5 |
Hb_000310_060 |
0.152905363 |
- |
- |
hypothetical protein JCGZ_20793 [Jatropha curcas] |
| 6 |
Hb_000310_120 |
0.1583299051 |
- |
- |
PREDICTED: probable transaldolase [Jatropha curcas] |
| 7 |
Hb_001587_040 |
0.1590802999 |
- |
- |
PREDICTED: soluble inorganic pyrophosphatase 6, chloroplastic [Jatropha curcas] |
| 8 |
Hb_001377_530 |
0.1593404425 |
- |
- |
NC domain-containing family protein [Populus trichocarpa] |
| 9 |
Hb_004650_030 |
0.1641735383 |
- |
- |
PREDICTED: uncharacterized protein LOC105638888 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000189_130 |
0.1651618392 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATA, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000392_250 |
0.1675583152 |
- |
- |
PREDICTED: HVA22-like protein a [Jatropha curcas] |
| 12 |
Hb_000996_080 |
0.1703305645 |
- |
- |
PREDICTED: intracellular protein transport protein USO1 [Jatropha curcas] |
| 13 |
Hb_000049_200 |
0.170460451 |
- |
- |
PREDICTED: uncharacterized protein LOC105644462 [Jatropha curcas] |
| 14 |
Hb_004195_140 |
0.1731975108 |
- |
- |
Sec14 cytosolic factor, putative [Ricinus communis] |
| 15 |
Hb_000270_280 |
0.1748827626 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 16 |
Hb_000689_050 |
0.1751808749 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001408_170 |
0.176419903 |
- |
- |
PREDICTED: phosphoglycolate phosphatase 2 [Jatropha curcas] |
| 18 |
Hb_004855_020 |
0.1782419159 |
- |
- |
hypothetical protein POPTR_0006s10010g [Populus trichocarpa] |
| 19 |
Hb_010868_040 |
0.1791559355 |
- |
- |
PREDICTED: reticulon-like protein B21 isoform X1 [Populus euphratica] |
| 20 |
Hb_005694_050 |
0.180130929 |
- |
- |
NC domain-containing family protein [Populus trichocarpa] |