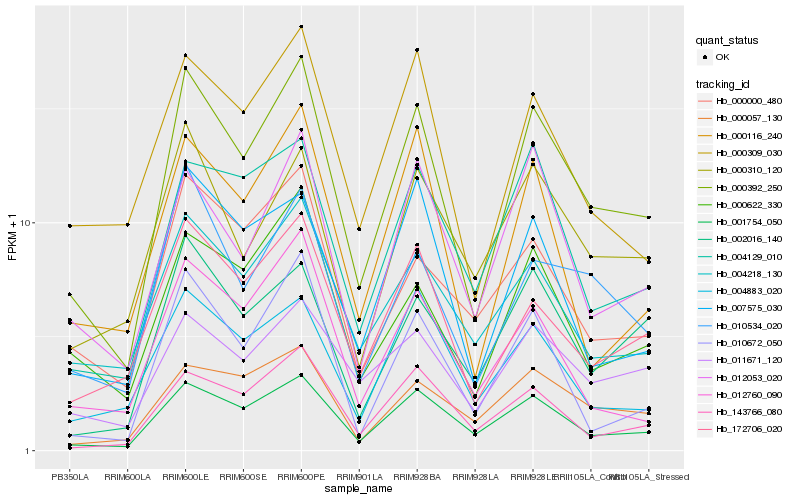
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000392_250 |
0.0 |
- |
- |
PREDICTED: HVA22-like protein a [Jatropha curcas] |
| 2 |
Hb_001754_050 |
0.0883377749 |
- |
- |
protein arginine n-methyltransferase, putative [Ricinus communis] |
| 3 |
Hb_010534_020 |
0.1211565316 |
- |
- |
PREDICTED: protein XRI1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_172706_020 |
0.1211832978 |
- |
- |
PREDICTED: protein ABIL2 [Jatropha curcas] |
| 5 |
Hb_000622_330 |
0.1236800936 |
- |
- |
PREDICTED: uncharacterized protein LOC105642906 [Jatropha curcas] |
| 6 |
Hb_012053_020 |
0.1277570452 |
- |
- |
PREDICTED: magnesium transporter MRS2-11, chloroplastic [Jatropha curcas] |
| 7 |
Hb_004218_130 |
0.1295639526 |
- |
- |
PREDICTED: uridine 5'-monophosphate synthase [Jatropha curcas] |
| 8 |
Hb_143766_080 |
0.1298426538 |
- |
- |
PREDICTED: uncharacterized protein LOC105637797 isoform X1 [Jatropha curcas] |
| 9 |
Hb_011671_120 |
0.1308112137 |
- |
- |
hypothetical protein JCGZ_26275 [Jatropha curcas] |
| 10 |
Hb_000309_030 |
0.1309744313 |
- |
- |
PREDICTED: formate dehydrogenase, mitochondrial [Jatropha curcas] |
| 11 |
Hb_000310_120 |
0.1336816563 |
- |
- |
PREDICTED: probable transaldolase [Jatropha curcas] |
| 12 |
Hb_000057_130 |
0.1347540675 |
- |
- |
PREDICTED: pyridoxal 5'-phosphate synthase-like subunit PDX1.2 [Jatropha curcas] |
| 13 |
Hb_007575_030 |
0.1366753172 |
- |
- |
PREDICTED: protein RCC2 homolog [Jatropha curcas] |
| 14 |
Hb_004129_010 |
0.1369016995 |
- |
- |
aspartate aminotransferase, putative [Ricinus communis] |
| 15 |
Hb_002016_140 |
0.1372500088 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase BAH1-like 1 [Jatropha curcas] |
| 16 |
Hb_000116_240 |
0.1391940955 |
transcription factor |
TF Family: HB |
homeobox-leucine zipper family protein [Populus trichocarpa] |
| 17 |
Hb_000000_480 |
0.1401007925 |
- |
- |
PREDICTED: deSI-like protein At4g17486 isoform X1 [Jatropha curcas] |
| 18 |
Hb_010672_050 |
0.1402879796 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 2 isoform X3 [Jatropha curcas] |
| 19 |
Hb_012760_090 |
0.1427945196 |
- |
- |
Aspartic proteinase-like protein 1 [Morus notabilis] |
| 20 |
Hb_004883_020 |
0.1429335577 |
- |
- |
PREDICTED: uncharacterized protein LOC105647351 [Jatropha curcas] |