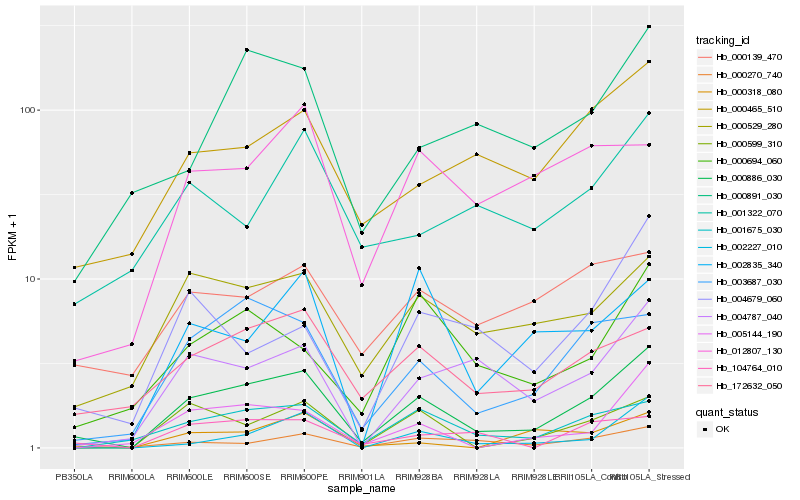
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000886_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105113410 [Populus euphratica] |
| 2 |
Hb_003687_030 |
0.185554369 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 3 |
Hb_001675_030 |
0.188744868 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000529_280 |
0.2009739491 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX1, chloroplastic [Jatropha curcas] |
| 5 |
Hb_172632_050 |
0.2052001631 |
- |
- |
PREDICTED: uncharacterized protein LOC105646134 [Jatropha curcas] |
| 6 |
Hb_004787_040 |
0.2056705969 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000465_510 |
0.2110098706 |
- |
- |
PREDICTED: GTP-binding protein YPTM2 [Populus euphratica] |
| 8 |
Hb_000599_310 |
0.2116236582 |
- |
- |
PREDICTED: pleckstrin homology domain-containing family A member 8-like [Jatropha curcas] |
| 9 |
Hb_000270_740 |
0.2151984195 |
- |
- |
PREDICTED: probable inactive receptor kinase At2g26730 [Jatropha curcas] |
| 10 |
Hb_104764_010 |
0.2154395822 |
- |
- |
hypothetical protein JCGZ_16024 [Jatropha curcas] |
| 11 |
Hb_000694_060 |
0.2194491158 |
- |
- |
Rotenone-insensitive NADH-ubiquinone oxidoreductase, mitochondrial precursor, putative [Ricinus communis] |
| 12 |
Hb_000891_030 |
0.2217918385 |
- |
- |
PREDICTED: uncharacterized protein At4g22758 [Jatropha curcas] |
| 13 |
Hb_005144_190 |
0.2219686833 |
- |
- |
hypothetical protein F383_09558 [Gossypium arboreum] |
| 14 |
Hb_002227_010 |
0.2310393012 |
- |
- |
PREDICTED: 7-deoxyloganetin glucosyltransferase-like [Populus euphratica] |
| 15 |
Hb_012807_130 |
0.2333652795 |
- |
- |
PREDICTED: lactoylglutathione lyase [Jatropha curcas] |
| 16 |
Hb_000139_470 |
0.2336753305 |
- |
- |
Dual specificity protein phosphatase, putative [Ricinus communis] |
| 17 |
Hb_004679_060 |
0.2354238364 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002835_340 |
0.2381393282 |
- |
- |
PREDICTED: peroxisomal primary amine oxidase [Jatropha curcas] |
| 19 |
Hb_000318_080 |
0.2391302453 |
- |
- |
PREDICTED: la-related protein 6C [Jatropha curcas] |
| 20 |
Hb_001322_070 |
0.2391422161 |
- |
- |
protein with unknown function [Ricinus communis] |