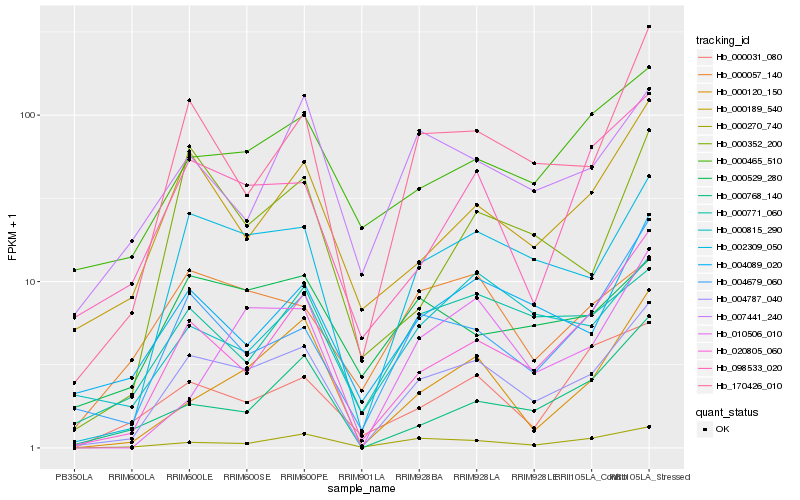
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004787_040 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002309_050 |
0.1041269873 |
- |
- |
beta-carotene hydroxylase, putative [Ricinus communis] |
| 3 |
Hb_170426_010 |
0.1392533972 |
- |
- |
cytochrome b5 isoform Cb5-B [Vernicia fordii] |
| 4 |
Hb_000270_740 |
0.1547649498 |
- |
- |
PREDICTED: probable inactive receptor kinase At2g26730 [Jatropha curcas] |
| 5 |
Hb_020805_060 |
0.1572360892 |
- |
- |
PREDICTED: protein S-acyltransferase 21 [Jatropha curcas] |
| 6 |
Hb_004679_060 |
0.1698430448 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000189_540 |
0.1715381919 |
- |
- |
PREDICTED: tRNA(adenine(34)) deaminase, chloroplastic [Jatropha curcas] |
| 8 |
Hb_004089_020 |
0.1738148139 |
- |
- |
PREDICTED: 3-phosphoinositide-dependent protein kinase 2 [Jatropha curcas] |
| 9 |
Hb_000120_150 |
0.1839330991 |
- |
- |
hypothetical protein JCGZ_08453 [Jatropha curcas] |
| 10 |
Hb_000771_060 |
0.1855633516 |
- |
- |
PREDICTED: spermine synthase [Jatropha curcas] |
| 11 |
Hb_000465_510 |
0.187343673 |
- |
- |
PREDICTED: GTP-binding protein YPTM2 [Populus euphratica] |
| 12 |
Hb_098533_020 |
0.1878661788 |
- |
- |
PREDICTED: zinc finger AN1 domain-containing stress-associated protein 12 [Jatropha curcas] |
| 13 |
Hb_000768_140 |
0.1925967289 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
tRNA (guanine-n(7)-)-methyltransferase, putative [Ricinus communis] |
| 14 |
Hb_000352_200 |
0.1967666139 |
- |
- |
PREDICTED: OTU domain-containing protein DDB_G0284757 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000815_290 |
0.1978105642 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X2 [Jatropha curcas] |
| 16 |
Hb_010506_010 |
0.1997134882 |
- |
- |
PREDICTED: 7-deoxyloganetin glucosyltransferase-like [Jatropha curcas] |
| 17 |
Hb_000057_140 |
0.2000255315 |
- |
- |
Mitochondrial carnitine/acylcarnitine carrier protein, putative [Ricinus communis] |
| 18 |
Hb_000031_080 |
0.200338436 |
- |
- |
PREDICTED: uncharacterized protein LOC105637482 [Jatropha curcas] |
| 19 |
Hb_000529_280 |
0.2010629476 |
- |
- |
PREDICTED: CBS domain-containing protein CBSX1, chloroplastic [Jatropha curcas] |
| 20 |
Hb_007441_240 |
0.2026388098 |
- |
- |
ethphon-induced protein [Hevea brasiliensis] |