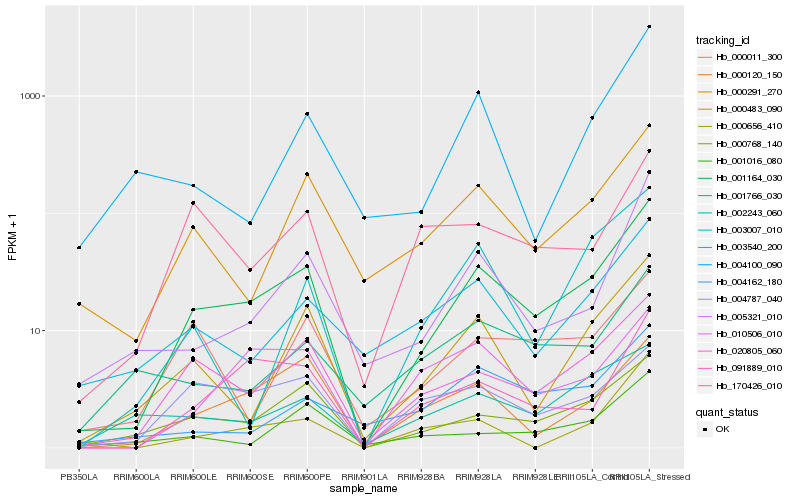
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001164_030 |
0.0 |
- |
- |
GMP synthase [glutamine-hydrolyzing] subunit A, putative [Ricinus communis] |
| 2 |
Hb_020805_060 |
0.1432957228 |
- |
- |
PREDICTED: protein S-acyltransferase 21 [Jatropha curcas] |
| 3 |
Hb_000768_140 |
0.1553712909 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
tRNA (guanine-n(7)-)-methyltransferase, putative [Ricinus communis] |
| 4 |
Hb_000483_090 |
0.177892021 |
- |
- |
- |
| 5 |
Hb_000291_270 |
0.1787849624 |
- |
- |
hypothetical protein JCGZ_12006 [Jatropha curcas] |
| 6 |
Hb_091889_010 |
0.187758755 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 7 |
Hb_000120_150 |
0.1881949146 |
- |
- |
hypothetical protein JCGZ_08453 [Jatropha curcas] |
| 8 |
Hb_003540_200 |
0.1981060077 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA3-like [Jatropha curcas] |
| 9 |
Hb_005321_010 |
0.2001833253 |
- |
- |
PREDICTED: uncharacterized protein LOC105644542 [Jatropha curcas] |
| 10 |
Hb_004787_040 |
0.2030155714 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_010506_010 |
0.208064196 |
- |
- |
PREDICTED: 7-deoxyloganetin glucosyltransferase-like [Jatropha curcas] |
| 12 |
Hb_002243_060 |
0.2091635486 |
- |
- |
hypothetical protein POPTR_0022s00750g [Populus trichocarpa] |
| 13 |
Hb_170426_010 |
0.2161138769 |
- |
- |
cytochrome b5 isoform Cb5-B [Vernicia fordii] |
| 14 |
Hb_001016_080 |
0.2172738408 |
- |
- |
hypothetical protein EUGRSUZ_J02452 [Eucalyptus grandis] |
| 15 |
Hb_000011_300 |
0.2174970081 |
- |
- |
PREDICTED: proteasome subunit alpha type-5 isoform X1 [Gossypium raimondii] |
| 16 |
Hb_003007_010 |
0.2186415872 |
- |
- |
PREDICTED: probable peptide/nitrate transporter At3g43790 isoform X1 [Populus euphratica] |
| 17 |
Hb_001766_030 |
0.2205756285 |
- |
- |
hypothetical protein POPTR_0017s12640g [Populus trichocarpa] |
| 18 |
Hb_004162_180 |
0.2235195761 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 19 |
Hb_000656_410 |
0.2243742417 |
- |
- |
hypothetical protein POPTR_0003s06410g [Populus trichocarpa] |
| 20 |
Hb_004100_090 |
0.2246168653 |
- |
- |
RecName: Full=Profilin-6; AltName: Full=Pollen allergen Hev b 8.0204; AltName: Allergen=Hev b 8.0204 [Hevea brasiliensis] |