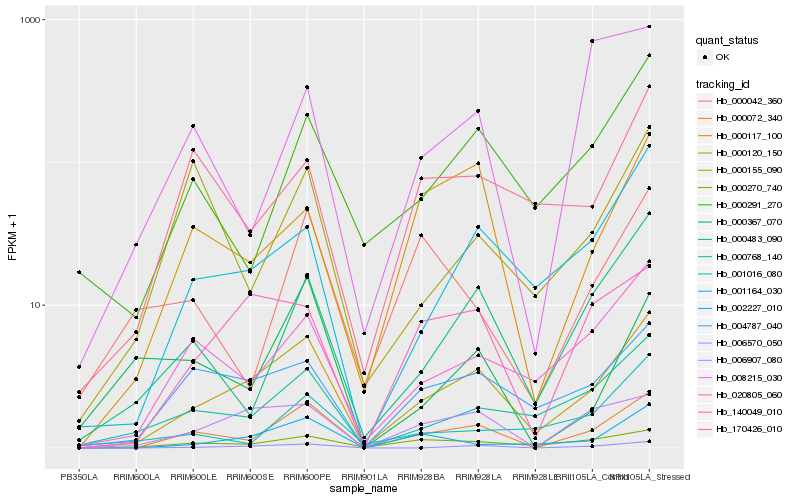
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000072_340 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000120_150 |
0.1588605617 |
- |
- |
hypothetical protein JCGZ_08453 [Jatropha curcas] |
| 3 |
Hb_000483_090 |
0.2085279253 |
- |
- |
- |
| 4 |
Hb_020805_060 |
0.2125603898 |
- |
- |
PREDICTED: protein S-acyltransferase 21 [Jatropha curcas] |
| 5 |
Hb_000270_740 |
0.2225589321 |
- |
- |
PREDICTED: probable inactive receptor kinase At2g26730 [Jatropha curcas] |
| 6 |
Hb_000291_270 |
0.2343156219 |
- |
- |
hypothetical protein JCGZ_12006 [Jatropha curcas] |
| 7 |
Hb_000768_140 |
0.236359751 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
tRNA (guanine-n(7)-)-methyltransferase, putative [Ricinus communis] |
| 8 |
Hb_006570_050 |
0.2369540991 |
- |
- |
PREDICTED: L-ascorbate oxidase homolog [Jatropha curcas] |
| 9 |
Hb_004787_040 |
0.2399843563 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000117_100 |
0.2425443037 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002227_010 |
0.2503253481 |
- |
- |
PREDICTED: 7-deoxyloganetin glucosyltransferase-like [Populus euphratica] |
| 12 |
Hb_001164_030 |
0.2515863087 |
- |
- |
GMP synthase [glutamine-hydrolyzing] subunit A, putative [Ricinus communis] |
| 13 |
Hb_006907_080 |
0.2522115141 |
- |
- |
PREDICTED: RPM1-interacting protein 4-like [Jatropha curcas] |
| 14 |
Hb_000155_090 |
0.2531272483 |
- |
- |
PREDICTED: glutaredoxin-C9 [Jatropha curcas] |
| 15 |
Hb_000042_360 |
0.2605408741 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_170426_010 |
0.2637600717 |
- |
- |
cytochrome b5 isoform Cb5-B [Vernicia fordii] |
| 17 |
Hb_008215_030 |
0.26922916 |
- |
- |
hypothetical protein 3 [Hevea brasiliensis] |
| 18 |
Hb_140049_010 |
0.270392652 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105632009 [Jatropha curcas] |
| 19 |
Hb_001016_080 |
0.2748717937 |
- |
- |
hypothetical protein EUGRSUZ_J02452 [Eucalyptus grandis] |
| 20 |
Hb_000367_070 |
0.2767980373 |
- |
- |
hypothetical protein JCGZ_11697 [Jatropha curcas] |