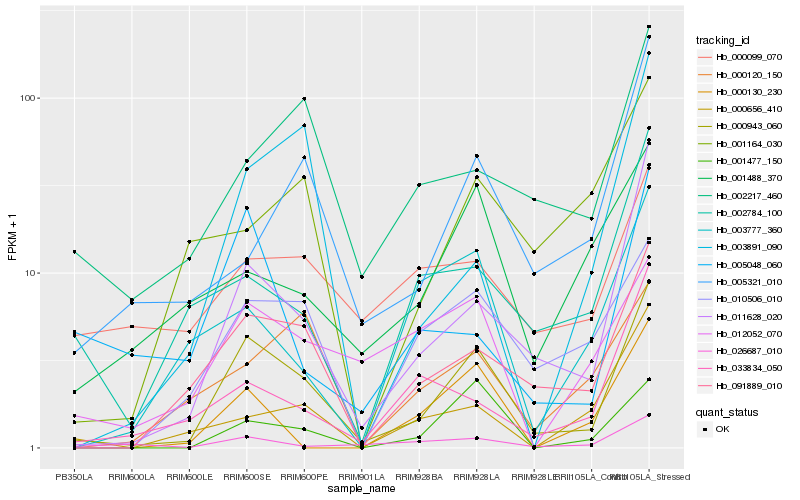
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000943_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635589 [Jatropha curcas] |
| 2 |
Hb_091889_010 |
0.1979257748 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 3 |
Hb_011628_020 |
0.2027313675 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_010506_010 |
0.237601077 |
- |
- |
PREDICTED: 7-deoxyloganetin glucosyltransferase-like [Jatropha curcas] |
| 5 |
Hb_001477_150 |
0.248071058 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 6 |
Hb_026687_010 |
0.2569631685 |
- |
- |
PREDICTED: 7-deoxyloganetin glucosyltransferase-like [Eucalyptus grandis] |
| 7 |
Hb_002217_460 |
0.2655550901 |
- |
- |
PREDICTED: transcription factor bHLH147-like [Jatropha curcas] |
| 8 |
Hb_005321_010 |
0.2694994087 |
- |
- |
PREDICTED: uncharacterized protein LOC105644542 [Jatropha curcas] |
| 9 |
Hb_012052_070 |
0.26956616 |
- |
- |
PREDICTED: uncharacterized protein LOC105635597 [Jatropha curcas] |
| 10 |
Hb_002784_100 |
0.2769186607 |
- |
- |
PREDICTED: uncharacterized protein LOC105631246 [Jatropha curcas] |
| 11 |
Hb_000130_230 |
0.2780270522 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF098-like [Jatropha curcas] |
| 12 |
Hb_000120_150 |
0.28029405 |
- |
- |
hypothetical protein JCGZ_08453 [Jatropha curcas] |
| 13 |
Hb_005048_060 |
0.2804410283 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003891_090 |
0.282424831 |
- |
- |
- |
| 15 |
Hb_033834_050 |
0.2829275002 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000656_410 |
0.2834178356 |
- |
- |
hypothetical protein POPTR_0003s06410g [Populus trichocarpa] |
| 17 |
Hb_001488_370 |
0.2834267779 |
- |
- |
Esterase PIR7B, putative [Ricinus communis] |
| 18 |
Hb_000099_070 |
0.2864068168 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 [Jatropha curcas] |
| 19 |
Hb_003777_360 |
0.2871576141 |
transcription factor |
TF Family: ERF |
AP2 domain-containing transcription factor family protein [Populus trichocarpa] |
| 20 |
Hb_001164_030 |
0.2873101634 |
- |
- |
GMP synthase [glutamine-hydrolyzing] subunit A, putative [Ricinus communis] |