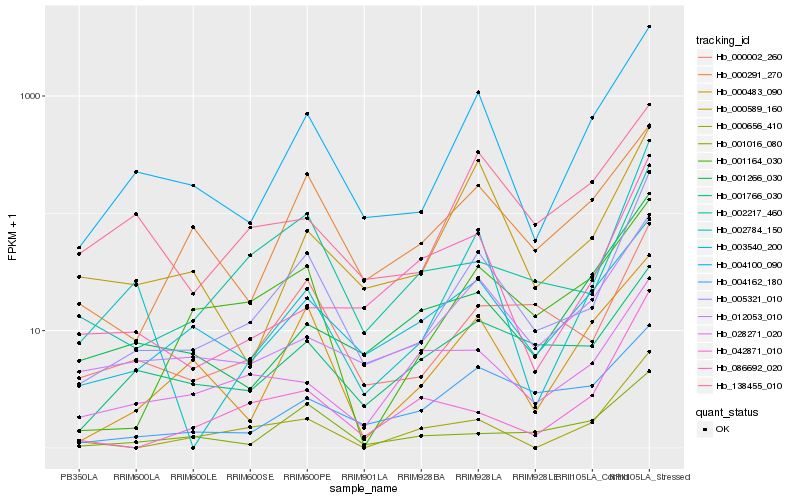
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005321_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644542 [Jatropha curcas] |
| 2 |
Hb_004100_090 |
0.1380429919 |
- |
- |
RecName: Full=Profilin-6; AltName: Full=Pollen allergen Hev b 8.0204; AltName: Allergen=Hev b 8.0204 [Hevea brasiliensis] |
| 3 |
Hb_000002_260 |
0.1660629454 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_012053_010 |
0.1838005532 |
- |
- |
srpk, putative [Ricinus communis] |
| 5 |
Hb_003540_200 |
0.1948892224 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA3-like [Jatropha curcas] |
| 6 |
Hb_002217_460 |
0.1950804313 |
- |
- |
PREDICTED: transcription factor bHLH147-like [Jatropha curcas] |
| 7 |
Hb_001164_030 |
0.2001833253 |
- |
- |
GMP synthase [glutamine-hydrolyzing] subunit A, putative [Ricinus communis] |
| 8 |
Hb_000291_270 |
0.2027354348 |
- |
- |
hypothetical protein JCGZ_12006 [Jatropha curcas] |
| 9 |
Hb_000483_090 |
0.2041508284 |
- |
- |
- |
| 10 |
Hb_000589_160 |
0.2075908203 |
- |
- |
PREDICTED: expansin-A4 [Jatropha curcas] |
| 11 |
Hb_000656_410 |
0.2102022392 |
- |
- |
hypothetical protein POPTR_0003s06410g [Populus trichocarpa] |
| 12 |
Hb_001766_030 |
0.2130479076 |
- |
- |
hypothetical protein POPTR_0017s12640g [Populus trichocarpa] |
| 13 |
Hb_001016_080 |
0.2137150412 |
- |
- |
hypothetical protein EUGRSUZ_J02452 [Eucalyptus grandis] |
| 14 |
Hb_138455_010 |
0.2162317161 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 15 |
Hb_042871_010 |
0.2177609456 |
- |
- |
PREDICTED: acyl-coenzyme A oxidase 3, peroxisomal-like [Jatropha curcas] |
| 16 |
Hb_086692_020 |
0.2182437529 |
- |
- |
PREDICTED: protein kinase APK1B, chloroplastic-like [Jatropha curcas] |
| 17 |
Hb_001266_030 |
0.2187854451 |
- |
- |
PREDICTED: lipid phosphate phosphatase epsilon 2, chloroplastic-like [Jatropha curcas] |
| 18 |
Hb_002784_150 |
0.2193692863 |
- |
- |
hypothetical protein POPTR_0001s06620g [Populus trichocarpa] |
| 19 |
Hb_004162_180 |
0.2209201347 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 20 |
Hb_028271_020 |
0.2309905312 |
- |
- |
acyl-CoA oxidase, putative [Ricinus communis] |