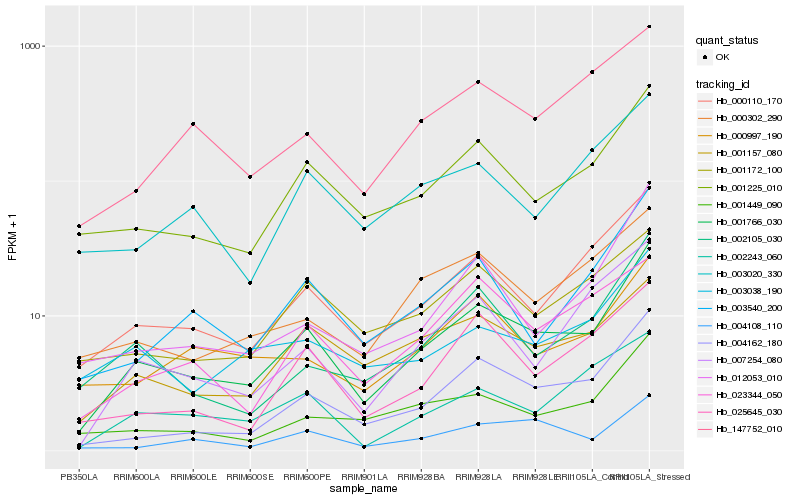
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001766_030 |
0.0 |
- |
- |
hypothetical protein POPTR_0017s12640g [Populus trichocarpa] |
| 2 |
Hb_004162_180 |
0.1239054341 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 3 |
Hb_000110_170 |
0.1272360238 |
- |
- |
PREDICTED: arginine biosynthesis bifunctional protein ArgJ, chloroplastic isoform X1 [Jatropha curcas] |
| 4 |
Hb_001225_010 |
0.1285043594 |
- |
- |
unnamed protein product [Coffea canephora] |
| 5 |
Hb_003540_200 |
0.1444606632 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA3-like [Jatropha curcas] |
| 6 |
Hb_001449_090 |
0.149674067 |
- |
- |
PREDICTED: NHL repeat-containing protein 2 isoform X2 [Jatropha curcas] |
| 7 |
Hb_002105_030 |
0.1546080581 |
- |
- |
- |
| 8 |
Hb_001157_080 |
0.1554965758 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 9 |
Hb_025645_030 |
0.1577277209 |
- |
- |
- |
| 10 |
Hb_007254_080 |
0.1582237074 |
- |
- |
- |
| 11 |
Hb_004108_110 |
0.1584872147 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_147752_010 |
0.1589358076 |
- |
- |
PREDICTED: 40S ribosomal protein S9-2-like isoform X1 [Populus euphratica] |
| 13 |
Hb_003038_190 |
0.1596214403 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105645961 [Jatropha curcas] |
| 14 |
Hb_012053_010 |
0.1605014777 |
- |
- |
srpk, putative [Ricinus communis] |
| 15 |
Hb_002243_060 |
0.1609771958 |
- |
- |
hypothetical protein POPTR_0022s00750g [Populus trichocarpa] |
| 16 |
Hb_000997_190 |
0.1620288437 |
- |
- |
PREDICTED: magnesium transporter MRS2-I-like [Jatropha curcas] |
| 17 |
Hb_000302_290 |
0.1625572094 |
- |
- |
40S ribosomal S10-3 -like protein [Gossypium arboreum] |
| 18 |
Hb_023344_050 |
0.1657595905 |
- |
- |
PREDICTED: uncharacterized protein LOC105644474 [Jatropha curcas] |
| 19 |
Hb_003020_330 |
0.1667028058 |
- |
- |
hypothetical protein B456_010G173100 [Gossypium raimondii] |
| 20 |
Hb_001172_100 |
0.1667855396 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex assembly factor 3-like isoform X1 [Jatropha curcas] |