| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
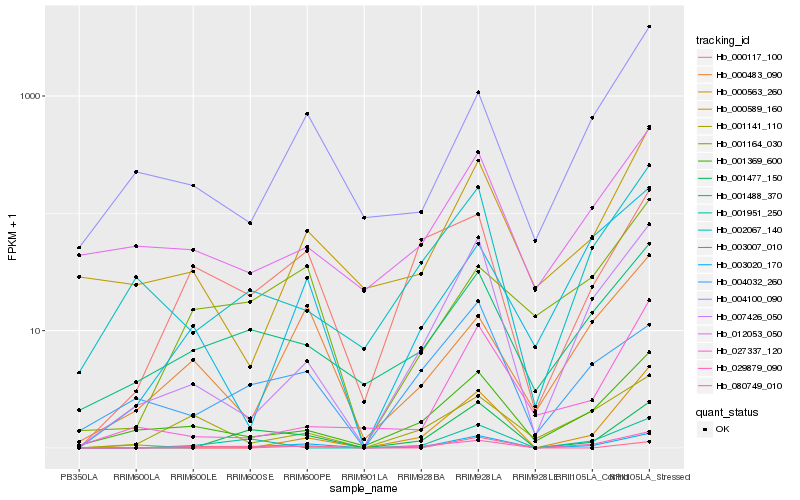
Hb_003020_170 |
0.0 |
- |
- |
Mitogen-activated protein kinase kinase kinase, putative [Ricinus communis] |
| 2 |
Hb_007426_050 |
0.2000573546 |
transcription factor |
TF Family: Tify |
JAZ8 [Hevea brasiliensis] |
| 3 |
Hb_001477_150 |
0.2237792156 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 4 |
Hb_001369_600 |
0.2261889444 |
- |
- |
kinase, putative [Ricinus communis] |
| 5 |
Hb_000117_100 |
0.2392327174 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001488_370 |
0.2457109875 |
- |
- |
Esterase PIR7B, putative [Ricinus communis] |
| 7 |
Hb_000589_160 |
0.247701695 |
- |
- |
PREDICTED: expansin-A4 [Jatropha curcas] |
| 8 |
Hb_029879_090 |
0.2493785419 |
- |
- |
PREDICTED: serine/threonine-protein kinase At3g07070 [Jatropha curcas] |
| 9 |
Hb_000563_260 |
0.2548393606 |
- |
- |
PREDICTED: uncharacterized protein LOC105641750 [Jatropha curcas] |
| 10 |
Hb_000483_090 |
0.2556210605 |
- |
- |
- |
| 11 |
Hb_001141_110 |
0.2596465301 |
- |
- |
hypothetical protein JCGZ_02206 [Jatropha curcas] |
| 12 |
Hb_080749_010 |
0.2621211721 |
- |
- |
PREDICTED: putative DNA-binding protein ESCAROLA [Populus euphratica] |
| 13 |
Hb_004100_090 |
0.2644550151 |
- |
- |
RecName: Full=Profilin-6; AltName: Full=Pollen allergen Hev b 8.0204; AltName: Allergen=Hev b 8.0204 [Hevea brasiliensis] |
| 14 |
Hb_002067_140 |
0.2648568769 |
- |
- |
- |
| 15 |
Hb_004032_260 |
0.2691853311 |
- |
- |
PREDICTED: uncharacterized protein LOC105635147 [Jatropha curcas] |
| 16 |
Hb_012053_050 |
0.2699614635 |
- |
- |
PREDICTED: UDP-D-apiose/UDP-D-xylose synthase 2 [Jatropha curcas] |
| 17 |
Hb_001164_030 |
0.2702513679 |
- |
- |
GMP synthase [glutamine-hydrolyzing] subunit A, putative [Ricinus communis] |
| 18 |
Hb_003007_010 |
0.2709506831 |
- |
- |
PREDICTED: probable peptide/nitrate transporter At3g43790 isoform X1 [Populus euphratica] |
| 19 |
Hb_001951_250 |
0.2727399004 |
- |
- |
- |
| 20 |
Hb_027337_120 |
0.2738627609 |
- |
- |
PREDICTED: uncharacterized protein LOC104430249 [Eucalyptus grandis] |