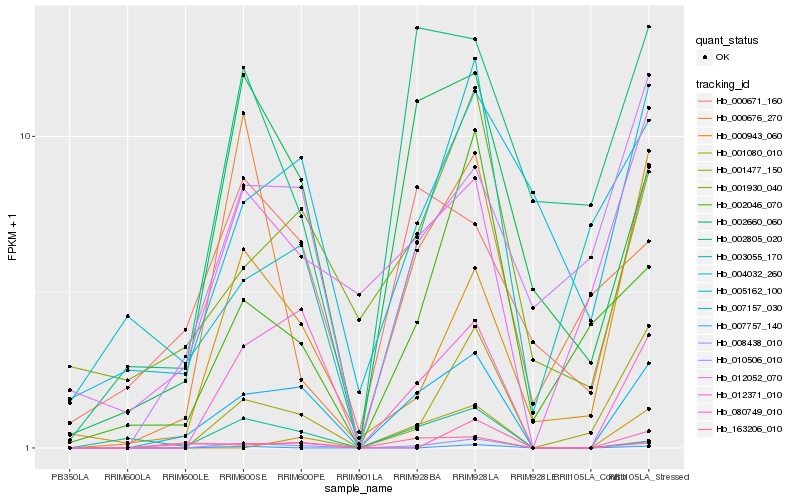
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008438_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104245771 [Nicotiana sylvestris] |
| 2 |
Hb_007157_030 |
0.1889410923 |
- |
- |
PREDICTED: protein LURP-one-related 6-like [Jatropha curcas] |
| 3 |
Hb_001477_150 |
0.2208621125 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 4 |
Hb_012371_010 |
0.2214039806 |
- |
- |
PREDICTED: uncharacterized protein LOC105641615 [Jatropha curcas] |
| 5 |
Hb_002660_060 |
0.2776359276 |
- |
- |
PREDICTED: uncharacterized protein LOC105642215 [Jatropha curcas] |
| 6 |
Hb_002046_070 |
0.2795662447 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-induced protein 22D-like isoform X1 [Populus euphratica] |
| 7 |
Hb_003055_170 |
0.3071534536 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor 25 isoform X1 [Jatropha curcas] |
| 8 |
Hb_007757_140 |
0.3122574533 |
- |
- |
PREDICTED: uncharacterized protein LOC104216022, partial [Nicotiana sylvestris] |
| 9 |
Hb_001930_040 |
0.3193901959 |
- |
- |
PREDICTED: serine acetyltransferase 1, chloroplastic-like [Beta vulgaris subsp. vulgaris] |
| 10 |
Hb_000676_270 |
0.322261856 |
transcription factor |
TF Family: SRS |
PREDICTED: protein SHI RELATED SEQUENCE 1 [Jatropha curcas] |
| 11 |
Hb_000943_060 |
0.3283945168 |
- |
- |
PREDICTED: uncharacterized protein LOC105635589 [Jatropha curcas] |
| 12 |
Hb_080749_010 |
0.3304340539 |
- |
- |
PREDICTED: putative DNA-binding protein ESCAROLA [Populus euphratica] |
| 13 |
Hb_002805_020 |
0.3397527973 |
transcription factor |
TF Family: SRS |
transcription factor, putative [Ricinus communis] |
| 14 |
Hb_004032_260 |
0.3413171531 |
- |
- |
PREDICTED: uncharacterized protein LOC105635147 [Jatropha curcas] |
| 15 |
Hb_012052_070 |
0.3499042321 |
- |
- |
PREDICTED: uncharacterized protein LOC105635597 [Jatropha curcas] |
| 16 |
Hb_005162_100 |
0.3500449698 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 17 |
Hb_010506_010 |
0.350678759 |
- |
- |
PREDICTED: 7-deoxyloganetin glucosyltransferase-like [Jatropha curcas] |
| 18 |
Hb_001080_010 |
0.3536662103 |
- |
- |
- |
| 19 |
Hb_000671_160 |
0.3573455459 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 20 |
Hb_163206_010 |
0.3602434586 |
- |
- |
hypothetical protein JCGZ_01203 [Jatropha curcas] |