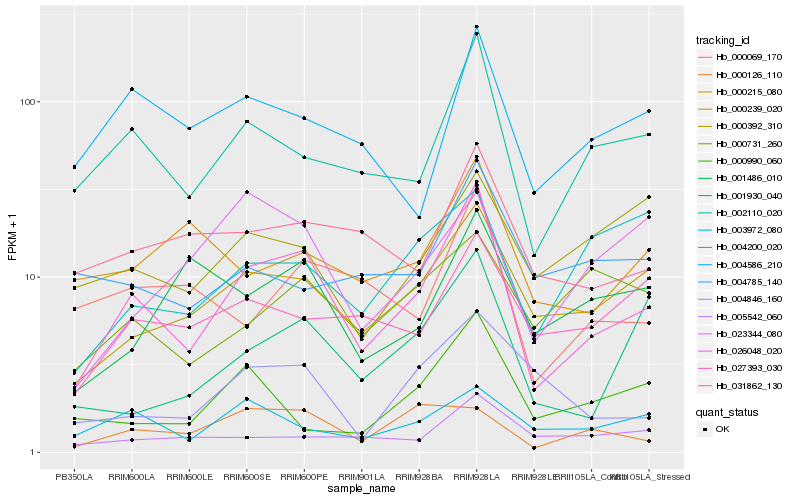
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_026048_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105645738 [Jatropha curcas] |
| 2 |
Hb_002110_020 |
0.1722767685 |
- |
- |
PREDICTED: transcription factor bHLH145 [Jatropha curcas] |
| 3 |
Hb_000239_020 |
0.1815772287 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At5g43190 [Jatropha curcas] |
| 4 |
Hb_031862_130 |
0.216200192 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 5 |
Hb_001930_040 |
0.2176831845 |
- |
- |
PREDICTED: serine acetyltransferase 1, chloroplastic-like [Beta vulgaris subsp. vulgaris] |
| 6 |
Hb_000069_170 |
0.2212222927 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 18 [Jatropha curcas] |
| 7 |
Hb_004586_210 |
0.2231331505 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 8 |
Hb_004846_160 |
0.2294313688 |
- |
- |
- |
| 9 |
Hb_027393_030 |
0.2298182573 |
- |
- |
PREDICTED: uncharacterized protein At5g39865 [Jatropha curcas] |
| 10 |
Hb_005542_060 |
0.232546955 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_004200_020 |
0.235224066 |
- |
- |
PREDICTED: uncharacterized protein LOC104878039 [Vitis vinifera] |
| 12 |
Hb_000215_080 |
0.2396419107 |
- |
- |
PREDICTED: uncharacterized protein LOC105637840 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000731_260 |
0.2417551378 |
- |
- |
hypothetical protein RCOM_0905090 [Ricinus communis] |
| 14 |
Hb_001486_010 |
0.2427564373 |
- |
- |
adenylsulfate kinase, putative [Ricinus communis] |
| 15 |
Hb_000126_110 |
0.2459084118 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase RFK1 isoform X3 [Populus euphratica] |
| 16 |
Hb_000990_060 |
0.2462814649 |
- |
- |
PREDICTED: protein EMBRYONIC FLOWER 1 [Jatropha curcas] |
| 17 |
Hb_004785_140 |
0.2465265722 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HERC2-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_023344_080 |
0.2468735696 |
- |
- |
PREDICTED: uncharacterized protein LOC105644472 isoform X1 [Jatropha curcas] |
| 19 |
Hb_003972_080 |
0.2511060384 |
- |
- |
PREDICTED: probable 6-phosphogluconolactonase 1 [Jatropha curcas] |
| 20 |
Hb_000392_310 |
0.2513626793 |
- |
- |
conserved hypothetical protein [Ricinus communis] |