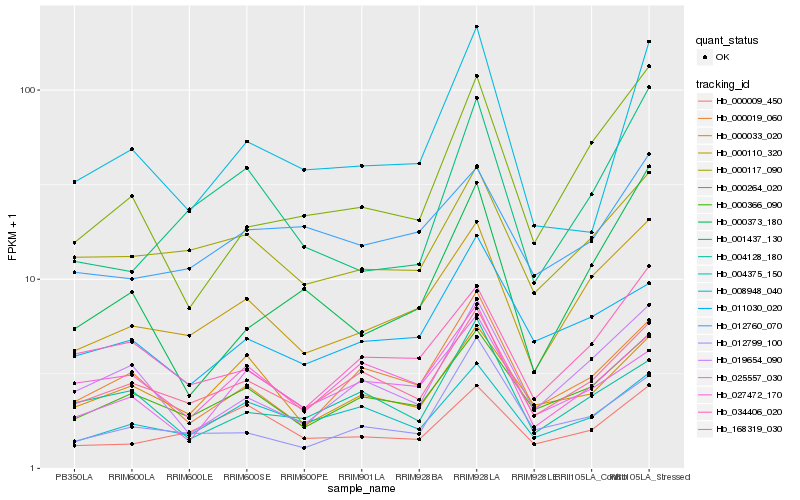
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008948_040 |
0.0 |
- |
- |
grr1, plant, putative [Ricinus communis] |
| 2 |
Hb_168319_030 |
0.1483492744 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g66520-like [Jatropha curcas] |
| 3 |
Hb_000019_060 |
0.1534985849 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g26782, mitochondrial [Jatropha curcas] |
| 4 |
Hb_000366_090 |
0.1649501228 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g32430, mitochondrial [Jatropha curcas] |
| 5 |
Hb_004375_150 |
0.1666208549 |
- |
- |
PREDICTED: uncharacterized protein LOC105642574 [Jatropha curcas] |
| 6 |
Hb_012799_100 |
0.1669905544 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g29230-like [Jatropha curcas] |
| 7 |
Hb_027472_170 |
0.1675589049 |
- |
- |
hypothetical protein POPTR_0007s08080g [Populus trichocarpa] |
| 8 |
Hb_004128_180 |
0.1701851426 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 9 |
Hb_000373_180 |
0.1742848042 |
transcription factor |
TF Family: Trihelix |
hypothetical protein JCGZ_25182 [Jatropha curcas] |
| 10 |
Hb_000110_320 |
0.1757213804 |
- |
- |
PREDICTED: ceramide-1-phosphate transfer protein [Jatropha curcas] |
| 11 |
Hb_034406_020 |
0.1772248223 |
- |
- |
PREDICTED: jmjC domain-containing protein 7 [Jatropha curcas] |
| 12 |
Hb_019654_090 |
0.1784725992 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g33680 [Jatropha curcas] |
| 13 |
Hb_025557_030 |
0.1786039351 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g74600, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000033_020 |
0.1790379861 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 15 |
Hb_000117_090 |
0.1797748441 |
- |
- |
PREDICTED: uncharacterized protein LOC105633456 [Jatropha curcas] |
| 16 |
Hb_012760_070 |
0.1813842793 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 17 |
Hb_001437_130 |
0.1822521854 |
- |
- |
hypothetical protein F383_15788 [Gossypium arboreum] |
| 18 |
Hb_000009_450 |
0.1837781741 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g22410, mitochondrial-like [Jatropha curcas] |
| 19 |
Hb_011030_020 |
0.185029884 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g21090 [Jatropha curcas] |
| 20 |
Hb_000264_020 |
0.1861671947 |
transcription factor |
TF Family: C2C2-Dof |
zinc finger protein, putative [Ricinus communis] |