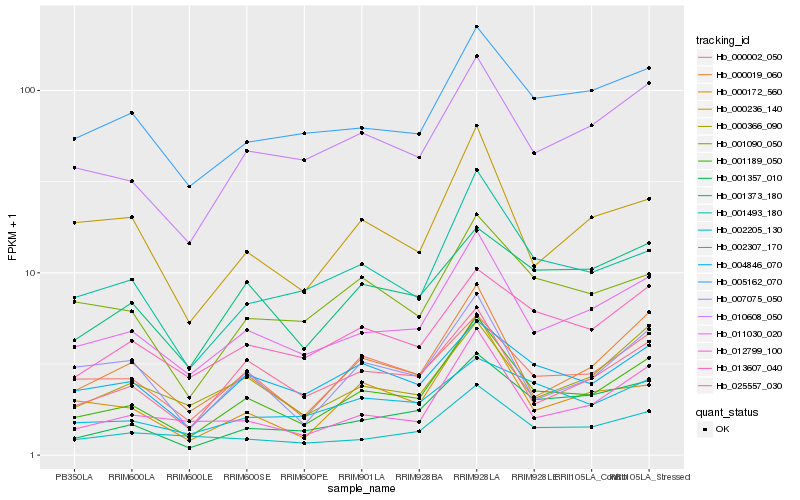
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001189_050 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g01570 [Jatropha curcas] |
| 2 |
Hb_011030_020 |
0.0793248264 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g21090 [Jatropha curcas] |
| 3 |
Hb_000019_060 |
0.0997808865 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g26782, mitochondrial [Jatropha curcas] |
| 4 |
Hb_001493_180 |
0.1062027226 |
- |
- |
fyve finger-containing phosphoinositide kinase, fyv1, putative [Ricinus communis] |
| 5 |
Hb_010608_050 |
0.1069801201 |
- |
- |
PREDICTED: uncharacterized protein LOC105645728 [Jatropha curcas] |
| 6 |
Hb_025557_030 |
0.1119809509 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g74600, chloroplastic [Jatropha curcas] |
| 7 |
Hb_001357_010 |
0.112357625 |
- |
- |
- |
| 8 |
Hb_000002_050 |
0.1148557835 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g33990 [Jatropha curcas] |
| 9 |
Hb_012799_100 |
0.1186880663 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g29230-like [Jatropha curcas] |
| 10 |
Hb_004846_070 |
0.1194101202 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g14730 [Populus euphratica] |
| 11 |
Hb_000172_560 |
0.1213768802 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02750 [Jatropha curcas] |
| 12 |
Hb_002205_130 |
0.1226282052 |
- |
- |
hypothetical protein B456_008G176100 [Gossypium raimondii] |
| 13 |
Hb_013607_040 |
0.1240177352 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g18520-like [Jatropha curcas] |
| 14 |
Hb_001090_050 |
0.1244533822 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_005162_070 |
0.1254234029 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001373_180 |
0.1267969294 |
- |
- |
PREDICTED: uncharacterized protein LOC105628780 isoform X2 [Jatropha curcas] |
| 17 |
Hb_007075_050 |
0.1277563843 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 18 |
Hb_000236_140 |
0.1284827446 |
- |
- |
PREDICTED: splicing regulatory glutamine/lysine-rich protein 1-like [Jatropha curcas] |
| 19 |
Hb_002307_170 |
0.130675885 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02750-like [Jatropha curcas] |
| 20 |
Hb_000366_090 |
0.1320148962 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g32430, mitochondrial [Jatropha curcas] |