| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
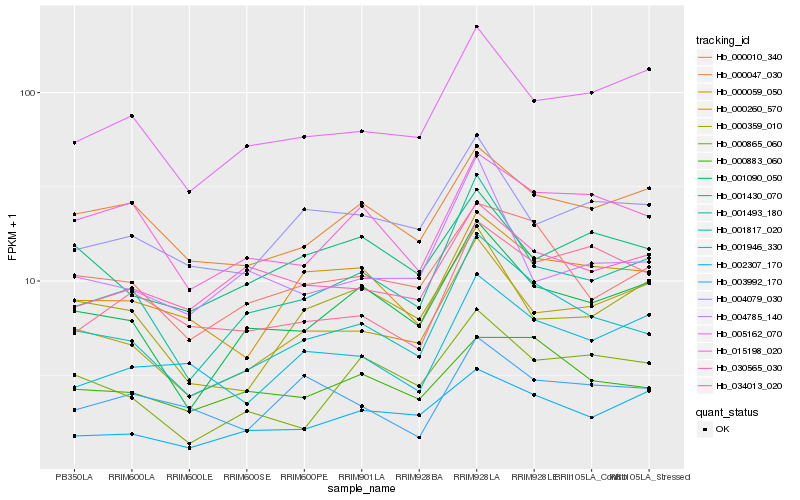
Hb_001817_020 |
0.0 |
- |
- |
PREDICTED: F-box/kelch-repeat protein OR23 [Jatropha curcas] |
| 2 |
Hb_001493_180 |
0.098763097 |
- |
- |
fyve finger-containing phosphoinositide kinase, fyv1, putative [Ricinus communis] |
| 3 |
Hb_001090_050 |
0.1034070207 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000010_340 |
0.1052525625 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g50280, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000059_050 |
0.1195695453 |
- |
- |
hypothetical protein JCGZ_25266 [Jatropha curcas] |
| 6 |
Hb_000260_570 |
0.1200645941 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 7 |
Hb_015198_020 |
0.1239616663 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000883_060 |
0.1278586899 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SNF2 domain-containing protein CLASSY 3-like [Jatropha curcas] |
| 9 |
Hb_000047_030 |
0.1336502012 |
- |
- |
tfiif-alpha, putative [Ricinus communis] |
| 10 |
Hb_005162_070 |
0.1346024268 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000865_060 |
0.1356455112 |
transcription factor |
TF Family: FAR1 |
hypothetical protein CISIN_1g043648mg [Citrus sinensis] |
| 12 |
Hb_004079_030 |
0.136004097 |
- |
- |
PREDICTED: uncharacterized protein LOC105637701 isoform X2 [Jatropha curcas] |
| 13 |
Hb_002307_170 |
0.1454712852 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02750-like [Jatropha curcas] |
| 14 |
Hb_004785_140 |
0.1471221867 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HERC2-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_001946_330 |
0.1472869312 |
- |
- |
PREDICTED: transmembrane protein 128 [Jatropha curcas] |
| 16 |
Hb_001430_070 |
0.1479940739 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_030565_030 |
0.148077108 |
- |
- |
PREDICTED: splicing regulatory glutamine/lysine-rich protein 1-like [Jatropha curcas] |
| 18 |
Hb_034013_020 |
0.1497988421 |
- |
- |
PREDICTED: protein trichome birefringence-like 11 [Jatropha curcas] |
| 19 |
Hb_000359_010 |
0.1520012724 |
- |
- |
o-linked n-acetylglucosamine transferase, ogt, putative [Ricinus communis] |
| 20 |
Hb_003992_170 |
0.1524985366 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 14-like [Populus euphratica] |