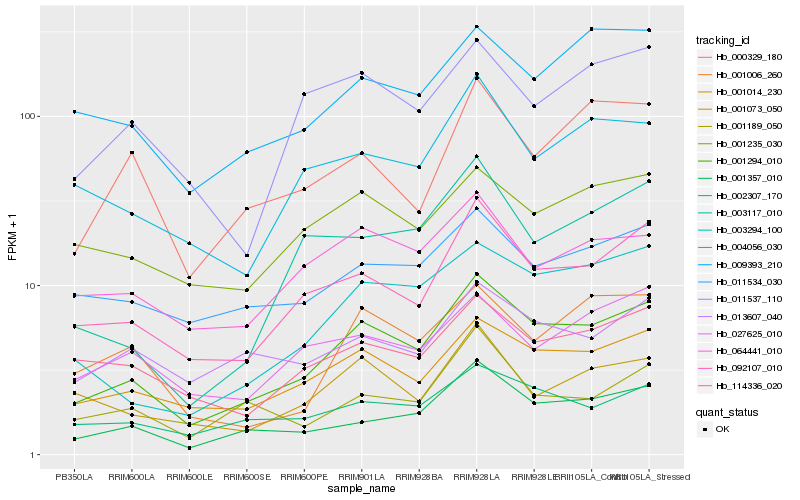
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001294_010 |
0.0 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF170-like [Jatropha curcas] |
| 2 |
Hb_092107_010 |
0.0947875115 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001014_230 |
0.09653703 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g11460 [Jatropha curcas] |
| 4 |
Hb_001357_010 |
0.1046948534 |
- |
- |
- |
| 5 |
Hb_003294_100 |
0.1162632401 |
- |
- |
hypothetical protein VITISV_009629 [Vitis vinifera] |
| 6 |
Hb_002307_170 |
0.1243465454 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02750-like [Jatropha curcas] |
| 7 |
Hb_004056_030 |
0.1296775299 |
- |
- |
hypothetical protein 33 [Hevea brasiliensis] |
| 8 |
Hb_064441_010 |
0.1344971357 |
- |
- |
PREDICTED: MACPF domain-containing protein CAD1-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_114336_020 |
0.1355338144 |
- |
- |
PREDICTED: cytosolic Fe-S cluster assembly factor narfl [Fragaria vesca subsp. vesca] |
| 10 |
Hb_009393_210 |
0.1357546776 |
- |
- |
PREDICTED: MHC class II regulatory factor RFX1-like [Jatropha curcas] |
| 11 |
Hb_011534_030 |
0.138223948 |
- |
- |
hypothetical protein POPTR_0008s08530g [Populus trichocarpa] |
| 12 |
Hb_001235_030 |
0.1396937078 |
- |
- |
PREDICTED: KH domain-containing protein At5g56140 [Vitis vinifera] |
| 13 |
Hb_011537_110 |
0.1405172969 |
- |
- |
hypothetical protein PRUPE_ppa013160mg [Prunus persica] |
| 14 |
Hb_001073_050 |
0.1405563394 |
- |
- |
- |
| 15 |
Hb_003117_010 |
0.1409225962 |
- |
- |
PREDICTED: uncharacterized protein LOC100259103 isoform X1 [Vitis vinifera] |
| 16 |
Hb_001006_260 |
0.1459378063 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit I-like [Jatropha curcas] |
| 17 |
Hb_000329_180 |
0.1460572794 |
- |
- |
Auxin-induced protein X10A, putative [Ricinus communis] |
| 18 |
Hb_027625_010 |
0.1467005042 |
- |
- |
PREDICTED: protein sym-1-like [Jatropha curcas] |
| 19 |
Hb_013607_040 |
0.1468111713 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g18520-like [Jatropha curcas] |
| 20 |
Hb_001189_050 |
0.1482043547 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g01570 [Jatropha curcas] |