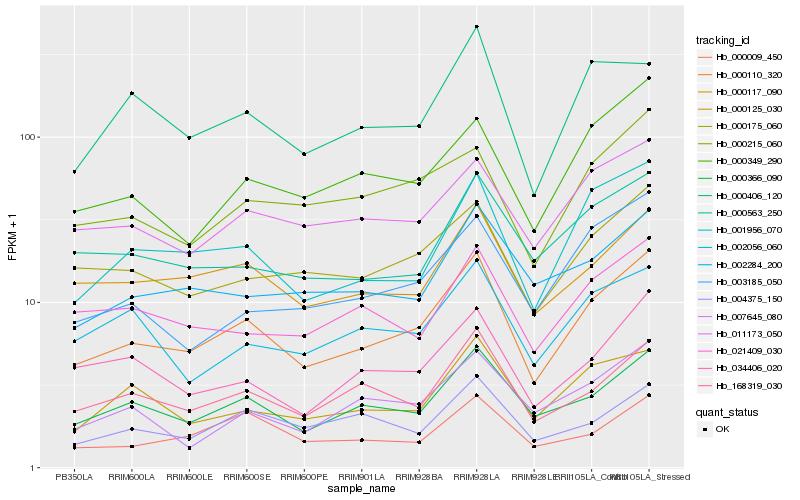
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000110_320 |
0.0 |
- |
- |
PREDICTED: ceramide-1-phosphate transfer protein [Jatropha curcas] |
| 2 |
Hb_000366_090 |
0.0897250393 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g32430, mitochondrial [Jatropha curcas] |
| 3 |
Hb_001956_070 |
0.0950416738 |
- |
- |
PREDICTED: telomere repeat-binding protein 2 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000117_090 |
0.1013116662 |
- |
- |
PREDICTED: uncharacterized protein LOC105633456 [Jatropha curcas] |
| 5 |
Hb_168319_030 |
0.1050571087 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g66520-like [Jatropha curcas] |
| 6 |
Hb_000175_060 |
0.1052607385 |
- |
- |
PREDICTED: uncharacterized protein LOC105634837 [Jatropha curcas] |
| 7 |
Hb_011173_050 |
0.1134376444 |
- |
- |
PREDICTED: OTU domain-containing protein DDB_G0284757 [Jatropha curcas] |
| 8 |
Hb_034406_020 |
0.1145101488 |
- |
- |
PREDICTED: jmjC domain-containing protein 7 [Jatropha curcas] |
| 9 |
Hb_000125_030 |
0.1159126838 |
transcription factor |
TF Family: mTERF |
hypothetical protein PRUPE_ppa019510mg, partial [Prunus persica] |
| 10 |
Hb_000349_290 |
0.1159529856 |
- |
- |
PREDICTED: zinc finger A20 and AN1 domain-containing stress-associated protein 1 [Jatropha curcas] |
| 11 |
Hb_000563_250 |
0.1163925689 |
- |
- |
PREDICTED: uncharacterized protein LOC105643472 [Jatropha curcas] |
| 12 |
Hb_000215_060 |
0.1176156719 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM17-2-like [Jatropha curcas] |
| 13 |
Hb_002284_200 |
0.119358212 |
- |
- |
PREDICTED: probable protein phosphatase 2C 27 isoform X1 [Jatropha curcas] |
| 14 |
Hb_003185_050 |
0.1205839764 |
- |
- |
PREDICTED: probable histone H2A variant 3 [Jatropha curcas] |
| 15 |
Hb_021409_030 |
0.1215789038 |
- |
- |
PREDICTED: serine/threonine-protein kinase WNK8 isoform X3 [Jatropha curcas] |
| 16 |
Hb_000009_450 |
0.1235842337 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g22410, mitochondrial-like [Jatropha curcas] |
| 17 |
Hb_000406_120 |
0.1239312113 |
- |
- |
acyl carrier protein 1, chloroplastic-like [Jatropha curcas] |
| 18 |
Hb_007645_080 |
0.1241942561 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g52630 [Jatropha curcas] |
| 19 |
Hb_002056_060 |
0.1245802451 |
- |
- |
Protein kinase APK1A, chloroplast precursor, putative [Ricinus communis] |
| 20 |
Hb_004375_150 |
0.1279702807 |
- |
- |
PREDICTED: uncharacterized protein LOC105642574 [Jatropha curcas] |