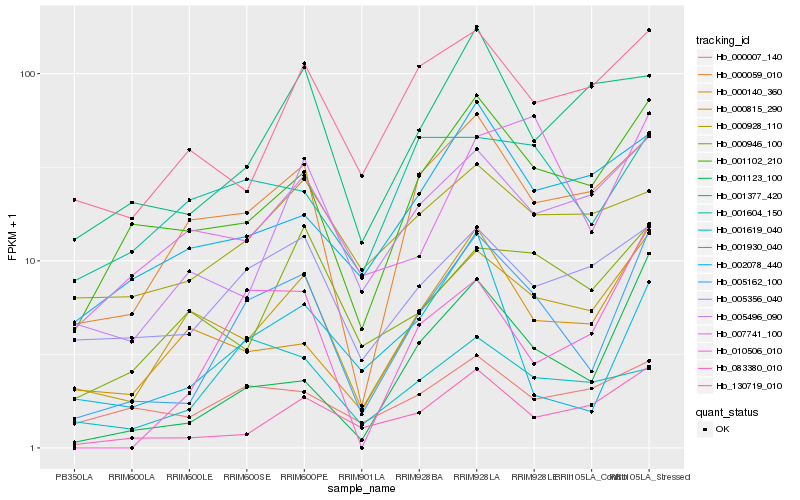
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005162_100 |
0.0 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 2 |
Hb_001102_210 |
0.1763285492 |
- |
- |
PREDICTED: alpha,alpha-trehalose-phosphate synthase [UDP-forming] 1-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_000059_010 |
0.1901872182 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 4 |
Hb_000815_290 |
0.1946509081 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X2 [Jatropha curcas] |
| 5 |
Hb_001123_100 |
0.1979282625 |
- |
- |
PREDICTED: uncharacterized protein LOC105647337 isoform X1 [Jatropha curcas] |
| 6 |
Hb_083380_010 |
0.206461653 |
- |
- |
- |
| 7 |
Hb_005356_040 |
0.2075472674 |
- |
- |
receptor serine-threonine protein kinase, putative [Ricinus communis] |
| 8 |
Hb_010506_010 |
0.217184319 |
- |
- |
PREDICTED: 7-deoxyloganetin glucosyltransferase-like [Jatropha curcas] |
| 9 |
Hb_005496_090 |
0.2175065104 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 1-like [Jatropha curcas] |
| 10 |
Hb_000140_360 |
0.2198763013 |
- |
- |
PREDICTED: uncharacterized protein LOC105642906 [Jatropha curcas] |
| 11 |
Hb_001930_040 |
0.2212507007 |
- |
- |
PREDICTED: serine acetyltransferase 1, chloroplastic-like [Beta vulgaris subsp. vulgaris] |
| 12 |
Hb_002078_440 |
0.2253346764 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 3 isoform X1 [Jatropha curcas] |
| 13 |
Hb_007741_100 |
0.2273332861 |
transcription factor |
TF Family: GRAS |
PREDICTED: LOW QUALITY PROTEIN: scarecrow-like protein 6 [Jatropha curcas] |
| 14 |
Hb_001377_420 |
0.2302420866 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_130719_010 |
0.2309290442 |
- |
- |
V-type proton ATPase subunit E [Hevea brasiliensis] |
| 16 |
Hb_000007_140 |
0.2323771893 |
- |
- |
PREDICTED: F-box/FBD/LRR-repeat protein At1g13570 [Jatropha curcas] |
| 17 |
Hb_001619_040 |
0.2341738323 |
- |
- |
PREDICTED: uncharacterized protein LOC105630601 [Jatropha curcas] |
| 18 |
Hb_000928_110 |
0.2371076588 |
- |
- |
PREDICTED: uncharacterized protein LOC105632499 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000946_100 |
0.2375640612 |
- |
- |
PREDICTED: SKI/DACH domain-containing protein 1-like [Jatropha curcas] |
| 20 |
Hb_001604_150 |
0.2403311026 |
- |
- |
PREDICTED: uncharacterized protein At3g15000, mitochondrial-like [Nelumbo nucifera] |