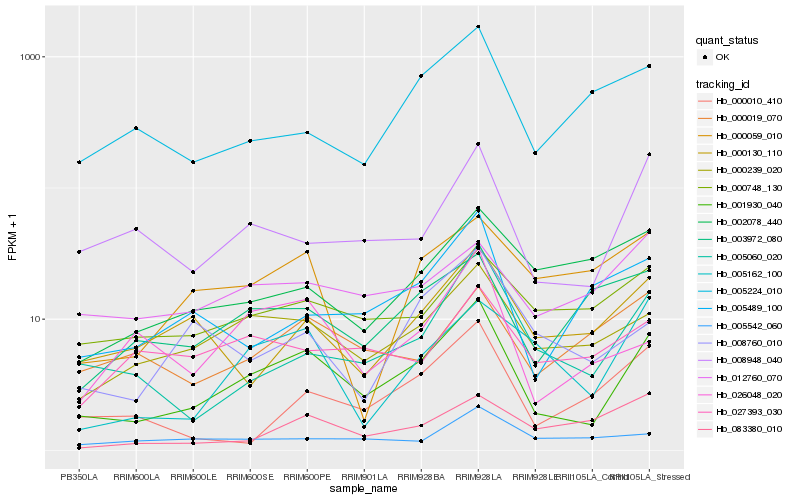
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001930_040 |
0.0 |
- |
- |
PREDICTED: serine acetyltransferase 1, chloroplastic-like [Beta vulgaris subsp. vulgaris] |
| 2 |
Hb_000239_020 |
0.1854662505 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At5g43190 [Jatropha curcas] |
| 3 |
Hb_008948_040 |
0.198285559 |
- |
- |
grr1, plant, putative [Ricinus communis] |
| 4 |
Hb_005060_020 |
0.2025456927 |
- |
- |
PREDICTED: metal tolerance protein 10-like isoform X1 [Populus euphratica] |
| 5 |
Hb_026048_020 |
0.2176831845 |
- |
- |
PREDICTED: uncharacterized protein LOC105645738 [Jatropha curcas] |
| 6 |
Hb_005162_100 |
0.2212507007 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 7 |
Hb_005489_100 |
0.2234536935 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000130_110 |
0.2243073399 |
- |
- |
PREDICTED: TPR repeat-containing thioredoxin TTL1 [Jatropha curcas] |
| 9 |
Hb_000748_130 |
0.2243792152 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein RICESLEEPER 2-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_000010_410 |
0.2294931189 |
- |
- |
hypothetical protein POPTR_0015s10070g, partial [Populus trichocarpa] |
| 11 |
Hb_003972_080 |
0.2311415 |
- |
- |
PREDICTED: probable 6-phosphogluconolactonase 1 [Jatropha curcas] |
| 12 |
Hb_008760_010 |
0.2319667405 |
- |
- |
berberine bridge enzyme [Hevea brasiliensis] |
| 13 |
Hb_005224_010 |
0.2333127498 |
- |
- |
glycine-rich protein [Bruguiera gymnorhiza] |
| 14 |
Hb_012760_070 |
0.235232051 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 15 |
Hb_000019_070 |
0.2365804847 |
- |
- |
PREDICTED: uncharacterized protein LOC105630200 [Jatropha curcas] |
| 16 |
Hb_027393_030 |
0.2421082593 |
- |
- |
PREDICTED: uncharacterized protein At5g39865 [Jatropha curcas] |
| 17 |
Hb_005542_060 |
0.2457415503 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 18 |
Hb_083380_010 |
0.2458739579 |
- |
- |
- |
| 19 |
Hb_000059_010 |
0.2467091908 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 20 |
Hb_002078_440 |
0.2479123068 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 3 isoform X1 [Jatropha curcas] |