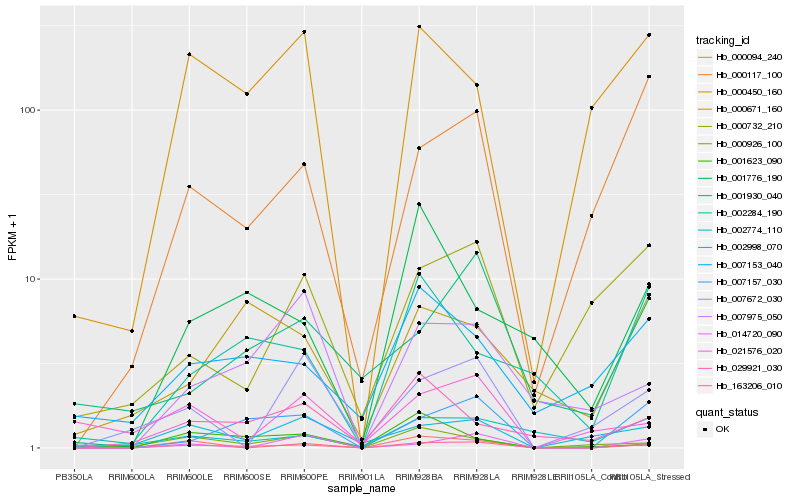
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_163206_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_01203 [Jatropha curcas] |
| 2 |
Hb_000094_240 |
0.2272813146 |
- |
- |
PREDICTED: uncharacterized protein LOC105633775 [Jatropha curcas] |
| 3 |
Hb_007157_030 |
0.2288241231 |
- |
- |
PREDICTED: protein LURP-one-related 6-like [Jatropha curcas] |
| 4 |
Hb_000450_160 |
0.2801019382 |
- |
- |
hypothetical protein JCGZ_16746 [Jatropha curcas] |
| 5 |
Hb_000926_100 |
0.2841873151 |
- |
- |
hypothetical protein B456_007G076800 [Gossypium raimondii] |
| 6 |
Hb_000117_100 |
0.2890783046 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002998_070 |
0.2939525935 |
- |
- |
hypothetical protein CARUB_v10019139mg, partial [Capsella rubella] |
| 8 |
Hb_014720_090 |
0.3042789721 |
- |
- |
- |
| 9 |
Hb_007153_040 |
0.3083372884 |
- |
- |
PREDICTED: uncharacterized protein LOC105633515 isoform X1 [Jatropha curcas] |
| 10 |
Hb_007975_050 |
0.3099131181 |
- |
- |
PREDICTED: uncharacterized protein LOC105649339 [Jatropha curcas] |
| 11 |
Hb_029921_030 |
0.3132725931 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_007672_030 |
0.3168713094 |
transcription factor |
TF Family: LOB |
LOB domain-containing 36 -like protein [Gossypium arboreum] |
| 13 |
Hb_002774_110 |
0.3212344152 |
- |
- |
heat shock protein binding protein, putative [Ricinus communis] |
| 14 |
Hb_002284_190 |
0.3243651869 |
- |
- |
PREDICTED: glutamate receptor 3.3 [Jatropha curcas] |
| 15 |
Hb_001776_190 |
0.3259332362 |
- |
- |
PREDICTED: protein kinase 2A, chloroplastic-like [Jatropha curcas] |
| 16 |
Hb_000671_160 |
0.3266771787 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001623_090 |
0.3274717668 |
- |
- |
PREDICTED: probable serine incorporator [Jatropha curcas] |
| 18 |
Hb_021576_020 |
0.3291667772 |
- |
- |
PREDICTED: uncharacterized protein LOC104426132 [Eucalyptus grandis] |
| 19 |
Hb_001930_040 |
0.330992404 |
- |
- |
PREDICTED: serine acetyltransferase 1, chloroplastic-like [Beta vulgaris subsp. vulgaris] |
| 20 |
Hb_000732_210 |
0.3318031069 |
- |
- |
hexose transporter 1 [Hevea brasiliensis] |