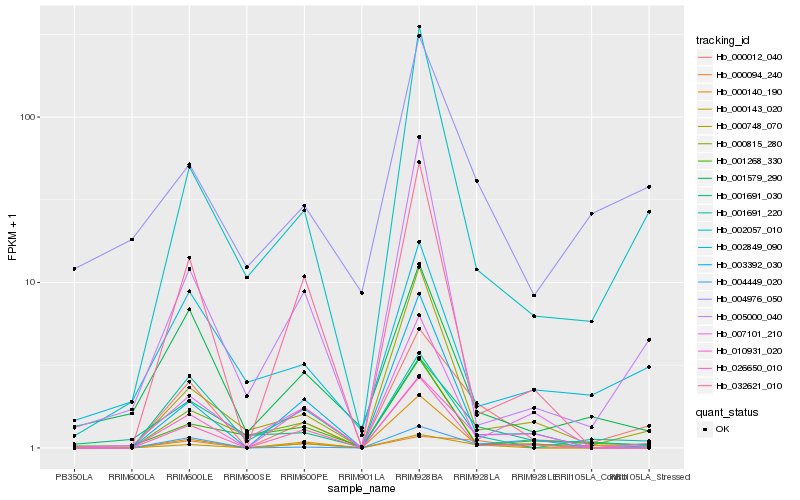
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000012_040 |
0.0 |
transcription factor |
TF Family: MYB-related |
PREDICTED: transcription factor RAX2-like [Populus euphratica] |
| 2 |
Hb_000140_190 |
0.2181794378 |
- |
- |
acc synthase, putative [Ricinus communis] |
| 3 |
Hb_002057_010 |
0.2347433692 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 4 |
Hb_001691_030 |
0.2391187764 |
- |
- |
lipoxygenase, putative [Ricinus communis] |
| 5 |
Hb_001691_220 |
0.244702496 |
- |
- |
PREDICTED: uncharacterized protein LOC105649537 [Jatropha curcas] |
| 6 |
Hb_000143_020 |
0.2509934283 |
- |
- |
PREDICTED: isoflavone 2'-hydroxylase-like [Jatropha curcas] |
| 7 |
Hb_000094_240 |
0.2535445583 |
- |
- |
PREDICTED: uncharacterized protein LOC105633775 [Jatropha curcas] |
| 8 |
Hb_001579_290 |
0.2612945946 |
- |
- |
PREDICTED: polygalacturonase At1g48100 isoform X1 [Jatropha curcas] |
| 9 |
Hb_004449_020 |
0.2664850448 |
- |
- |
PREDICTED: ATPase 10, plasma membrane-type isoform X3 [Elaeis guineensis] |
| 10 |
Hb_004976_050 |
0.2667047002 |
- |
- |
- |
| 11 |
Hb_000748_070 |
0.2715398756 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0005s22480g [Populus trichocarpa] |
| 12 |
Hb_026650_010 |
0.2745544024 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 13 |
Hb_002849_090 |
0.2816822839 |
- |
- |
hypothetical protein POPTR_0002s16380g [Populus trichocarpa] |
| 14 |
Hb_005000_040 |
0.2834543206 |
- |
- |
ACC oxidase ACCO2 [Manihot esculenta] |
| 15 |
Hb_032621_010 |
0.2901008235 |
- |
- |
hypothetical protein PHAVU_006G197300g [Phaseolus vulgaris] |
| 16 |
Hb_010931_020 |
0.2908891167 |
- |
- |
Kunitz-type protease inhibitor KPI-F4 [Populus trichocarpa x Populus deltoides] |
| 17 |
Hb_003392_030 |
0.2917835093 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_007101_210 |
0.2933572374 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 19 |
Hb_000815_280 |
0.2964476473 |
- |
- |
PREDICTED: phosphate transporter PHO1 homolog 3 [Jatropha curcas] |
| 20 |
Hb_001268_330 |
0.3060878219 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |